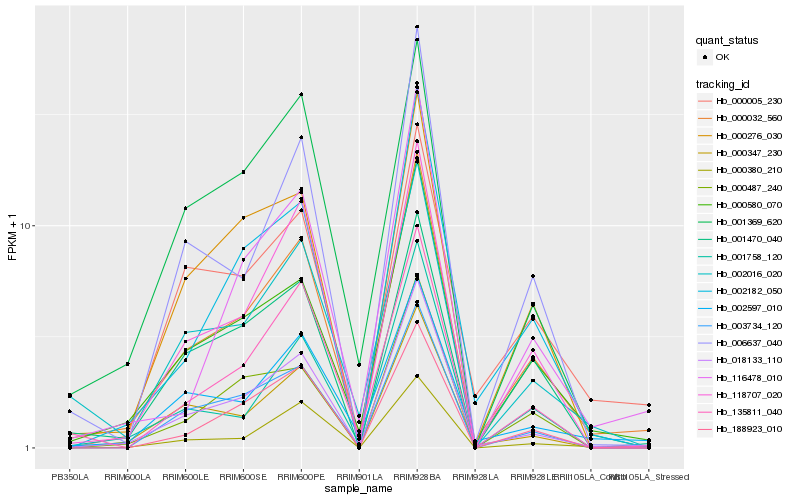
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_560 |
0.0 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 2 |
Hb_018133_110 |
0.072754146 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000580_070 |
0.0820116318 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 4 |
Hb_118707_020 |
0.1074583657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002597_010 |
0.1120032125 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 6 |
Hb_135811_040 |
0.1201021589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000276_030 |
0.1210907985 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_002182_050 |
0.1211671039 |
- |
- |
hypothetical protein CICLE_v10022654mg [Citrus clementina] |
| 9 |
Hb_002016_020 |
0.1215073037 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 10 |
Hb_000380_210 |
0.1224001687 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 11 |
Hb_000347_230 |
0.1235249467 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 12 |
Hb_001470_040 |
0.1252790745 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 13 |
Hb_006637_040 |
0.1254500524 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 14 |
Hb_003734_120 |
0.1264095532 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000005_230 |
0.12814678 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 16 |
Hb_001758_120 |
0.1321453284 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_116478_010 |
0.1321612965 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 18 |
Hb_000487_240 |
0.1333398825 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 19 |
Hb_001369_620 |
0.1342857605 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 20 |
Hb_188923_010 |
0.1369387474 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |