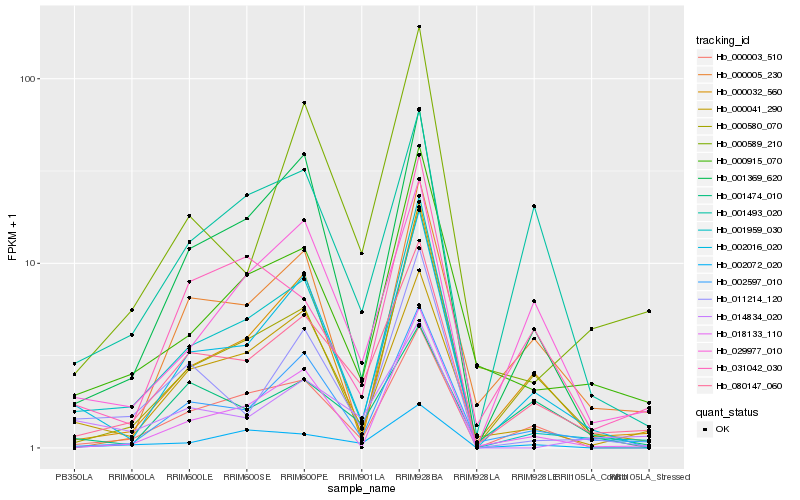
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_060 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002597_010 |
0.1289174319 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 3 |
Hb_000003_510 |
0.153527204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 4 |
Hb_000041_290 |
0.1598651913 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002016_020 |
0.1605113997 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 6 |
Hb_000580_070 |
0.1632448056 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 7 |
Hb_001959_030 |
0.1659500391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001474_010 |
0.1671966123 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000915_070 |
0.171831736 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001369_620 |
0.1735602185 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 11 |
Hb_000032_560 |
0.1747587629 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 12 |
Hb_029977_010 |
0.1786384487 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_000589_210 |
0.1801692644 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 14 |
Hb_000005_230 |
0.1811052241 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 15 |
Hb_002072_020 |
0.1848314158 |
- |
- |
hypothetical protein JCGZ_16057 [Jatropha curcas] |
| 16 |
Hb_031042_030 |
0.1871276847 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001493_020 |
0.188941071 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 18 |
Hb_011214_120 |
0.1913167738 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 19 |
Hb_014834_020 |
0.1931960612 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_018133_110 |
0.1970822248 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |