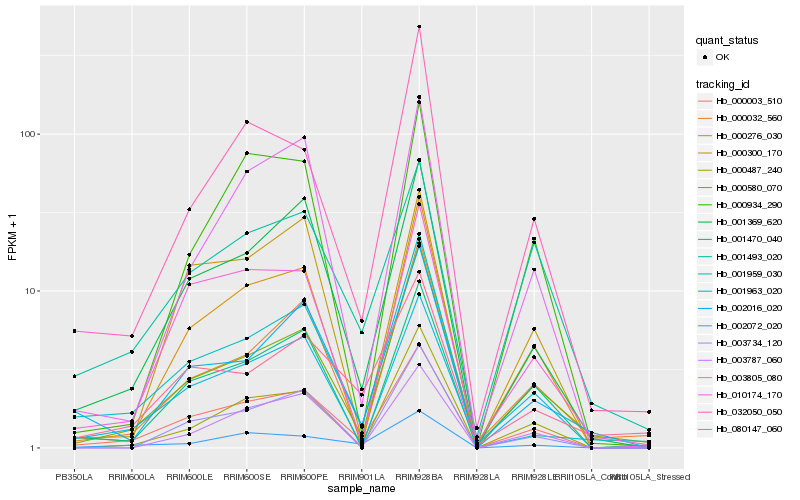
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_510 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 2 |
Hb_001963_020 |
0.0985253361 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001369_620 |
0.1001369413 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 4 |
Hb_000276_030 |
0.1153848527 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 5 |
Hb_001470_040 |
0.1217485123 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 6 |
Hb_002072_020 |
0.1266076792 |
- |
- |
hypothetical protein JCGZ_16057 [Jatropha curcas] |
| 7 |
Hb_010174_170 |
0.1279145045 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000580_070 |
0.1353283804 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 9 |
Hb_003734_120 |
0.1378694864 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002016_020 |
0.1428991755 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 11 |
Hb_001493_020 |
0.1486400338 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 12 |
Hb_000032_560 |
0.1490658577 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 13 |
Hb_003787_060 |
0.1490819555 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 14 |
Hb_001959_030 |
0.1494292463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003805_080 |
0.1514474846 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 16 |
Hb_080147_060 |
0.153527204 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000487_240 |
0.1538422463 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 18 |
Hb_032050_050 |
0.1568402807 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 19 |
Hb_000300_170 |
0.1572570155 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 20 |
Hb_000934_290 |
0.1586360417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |