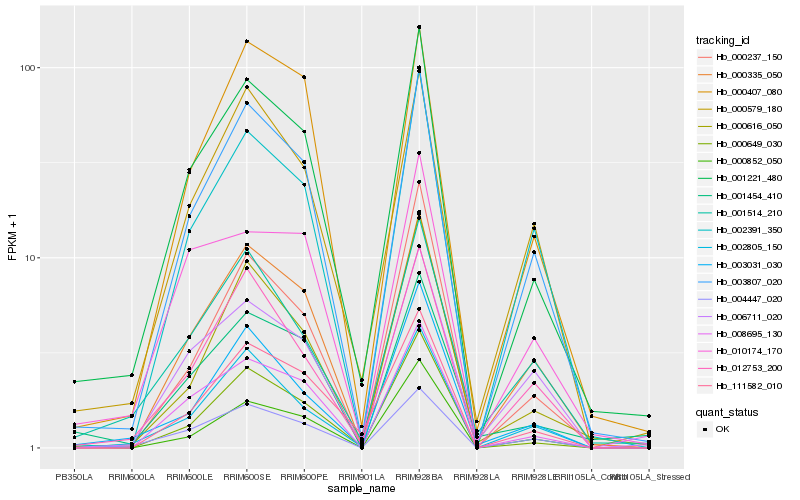
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_480 |
0.0 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_003807_020 |
0.0857864869 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 3 |
Hb_000649_030 |
0.10253721 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 4 |
Hb_004447_020 |
0.1083603203 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 5 |
Hb_111582_010 |
0.1131706484 |
- |
- |
hypothetical protein POPTR_0303s002201g, partial [Populus trichocarpa] |
| 6 |
Hb_000335_050 |
0.1138951618 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001454_410 |
0.1163382765 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 8 |
Hb_000852_050 |
0.1200200879 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 9 |
Hb_002805_150 |
0.1218623438 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_000616_050 |
0.1224820818 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 11 |
Hb_008695_130 |
0.1230988149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010174_170 |
0.1231387963 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000579_180 |
0.1267773778 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 14 |
Hb_012753_200 |
0.1275948486 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 15 |
Hb_003031_030 |
0.1276919894 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 16 |
Hb_000407_080 |
0.1280990788 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 17 |
Hb_006711_020 |
0.1293530993 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 18 |
Hb_000237_150 |
0.1300151576 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 19 |
Hb_001514_210 |
0.1306717469 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 20 |
Hb_002391_350 |
0.1309469331 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |