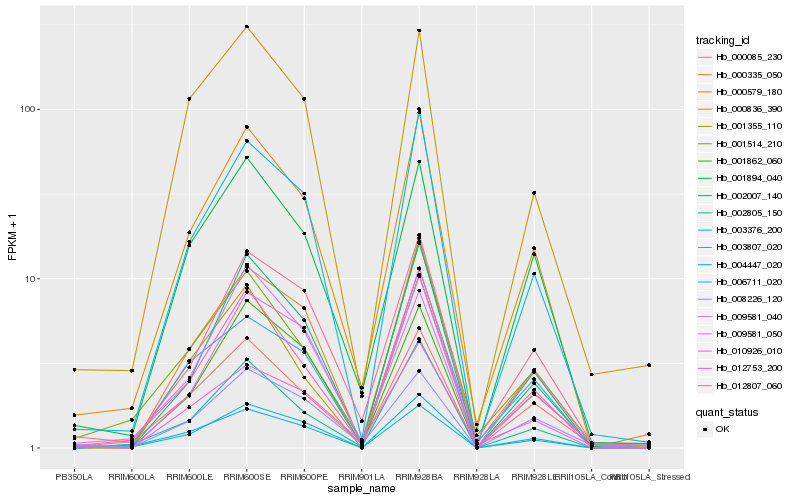
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_180 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 2 |
Hb_012753_200 |
0.0761263804 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 3 |
Hb_003807_020 |
0.0800262941 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 4 |
Hb_000085_230 |
0.0857043547 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 5 |
Hb_010926_010 |
0.0869280236 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 6 |
Hb_012807_060 |
0.0871506807 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_001894_040 |
0.0876438572 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000335_050 |
0.0900162737 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001514_210 |
0.0964877698 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 10 |
Hb_002805_150 |
0.0971454623 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_002007_140 |
0.0998028608 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 12 |
Hb_004447_020 |
0.1020579246 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 13 |
Hb_009581_050 |
0.1068036303 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 14 |
Hb_001862_060 |
0.1077030244 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003376_200 |
0.1085553213 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_006711_020 |
0.1133523436 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 17 |
Hb_000836_390 |
0.1143117248 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 18 |
Hb_008226_120 |
0.1163713796 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_009581_040 |
0.1228347767 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 20 |
Hb_001355_110 |
0.1233585706 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |