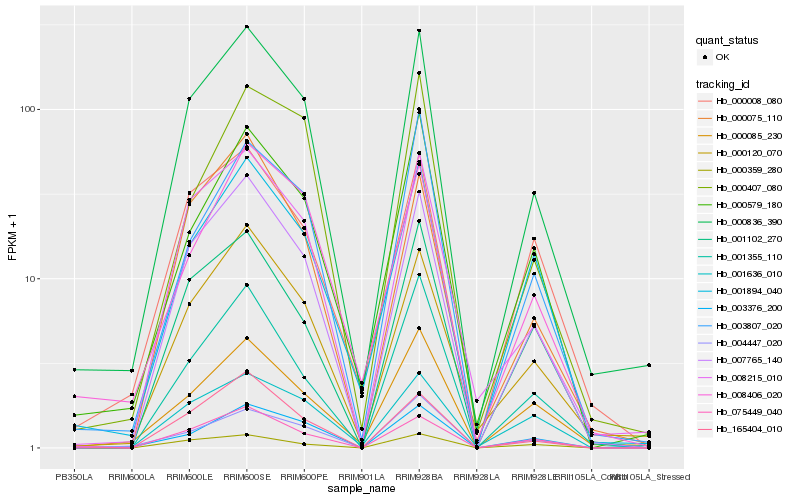
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_390 |
0.0 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 2 |
Hb_007765_140 |
0.0696103661 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 3 |
Hb_008215_010 |
0.0758656956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000120_070 |
0.0780198909 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 5 |
Hb_075449_040 |
0.1081371273 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 6 |
Hb_003376_200 |
0.1122449053 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000075_110 |
0.1133536942 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 8 |
Hb_001894_040 |
0.1143043236 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000579_180 |
0.1143117248 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 10 |
Hb_000085_230 |
0.1152723706 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 11 |
Hb_165404_010 |
0.1153771898 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 12 |
Hb_008406_020 |
0.1228306735 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 13 |
Hb_004447_020 |
0.123454873 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 14 |
Hb_001102_270 |
0.1258257262 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001636_010 |
0.1266857077 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 16 |
Hb_003807_020 |
0.1280947959 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 17 |
Hb_001355_110 |
0.1285094657 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000008_080 |
0.1287581566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000407_080 |
0.1339010742 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 20 |
Hb_000359_280 |
0.1340091553 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |