| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
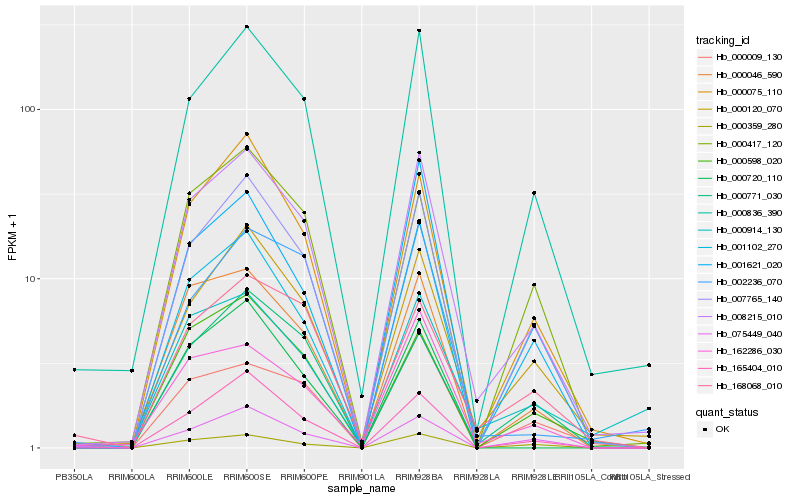
Hb_008215_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000836_390 |
0.0758656956 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 3 |
Hb_007765_140 |
0.0841215825 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 4 |
Hb_000120_070 |
0.0939600947 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 5 |
Hb_000075_110 |
0.1020624071 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_000046_590 |
0.104286723 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 7 |
Hb_165404_010 |
0.1091300415 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 8 |
Hb_162286_030 |
0.129834962 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 9 |
Hb_000598_020 |
0.1313854412 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 10 |
Hb_075449_040 |
0.1317301069 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 11 |
Hb_000359_280 |
0.1334106302 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 12 |
Hb_001102_270 |
0.1355213447 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_000914_130 |
0.137561089 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 14 |
Hb_000009_130 |
0.1408765887 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 15 |
Hb_000771_030 |
0.1412027144 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000417_120 |
0.1426779914 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 17 |
Hb_000720_110 |
0.1448327408 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 18 |
Hb_001621_020 |
0.1460286938 |
- |
- |
- |
| 19 |
Hb_168068_010 |
0.1479255226 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_002236_070 |
0.1494409683 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |