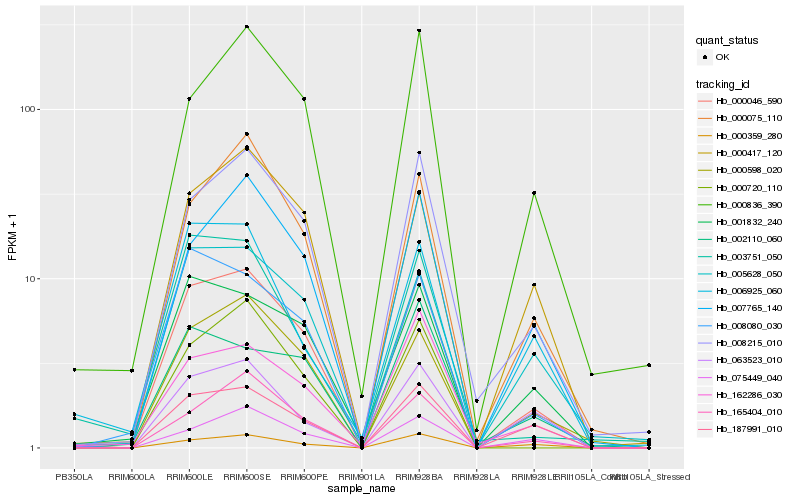
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_590 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_008215_010 |
0.104286723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001832_240 |
0.114696405 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 4 |
Hb_000598_020 |
0.1163747871 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 5 |
Hb_008080_030 |
0.124969875 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 6 |
Hb_007765_140 |
0.1280411319 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 7 |
Hb_187991_010 |
0.1285689894 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_005628_050 |
0.1290944596 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_000075_110 |
0.1319274872 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 10 |
Hb_000417_120 |
0.1354034289 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 11 |
Hb_000836_390 |
0.1357483059 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 12 |
Hb_165404_010 |
0.1362801293 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 13 |
Hb_000720_110 |
0.1383618563 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 14 |
Hb_162286_030 |
0.1399633369 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 15 |
Hb_000359_280 |
0.1403947307 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 16 |
Hb_075449_040 |
0.1519309831 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 17 |
Hb_003751_050 |
0.1548852561 |
- |
- |
PREDICTED: amino acid permease 4-like [Jatropha curcas] |
| 18 |
Hb_063523_010 |
0.1580506893 |
- |
- |
- |
| 19 |
Hb_002110_060 |
0.1605374521 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 20 |
Hb_006925_060 |
0.1611452805 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |