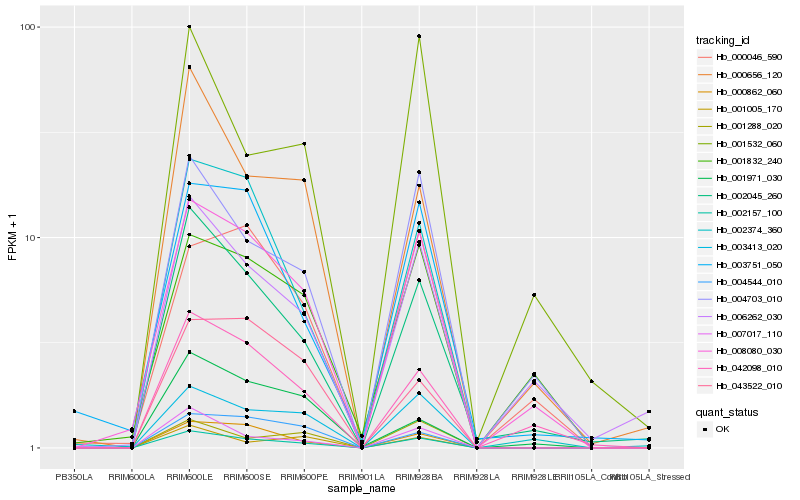
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_003413_020 |
0.1005894663 |
- |
- |
- |
| 3 |
Hb_000862_060 |
0.1145264258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002045_260 |
0.114610547 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 5 |
Hb_008080_030 |
0.1152157498 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 6 |
Hb_007017_110 |
0.1229336072 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_002374_360 |
0.131836809 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 8 |
Hb_004703_010 |
0.1465028332 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_000656_120 |
0.1586083668 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 10 |
Hb_042098_010 |
0.1608907045 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 11 |
Hb_006262_030 |
0.1612693346 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_004544_010 |
0.1654888029 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 13 |
Hb_001288_020 |
0.1681226611 |
- |
- |
- |
| 14 |
Hb_003751_050 |
0.1701495226 |
- |
- |
PREDICTED: amino acid permease 4-like [Jatropha curcas] |
| 15 |
Hb_001005_170 |
0.1707668501 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 16 |
Hb_043522_010 |
0.1850608176 |
- |
- |
- |
| 17 |
Hb_001832_240 |
0.1885005675 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 18 |
Hb_001532_060 |
0.1972884006 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000046_590 |
0.2001925906 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_001971_030 |
0.2016036137 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |