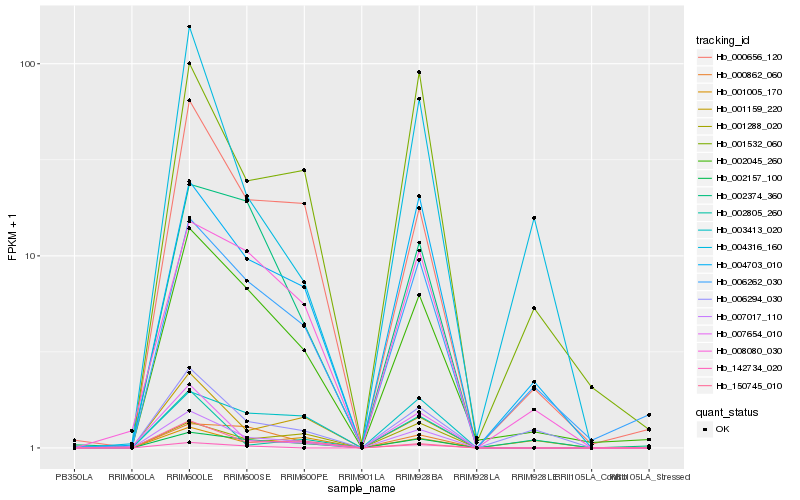
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007017_110 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 2 |
Hb_002157_100 |
0.1229336072 |
- |
- |
- |
| 3 |
Hb_002045_260 |
0.1406316974 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 4 |
Hb_000656_120 |
0.1588056056 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 5 |
Hb_004703_010 |
0.159215414 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 6 |
Hb_007654_010 |
0.1665356175 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 7 |
Hb_001005_170 |
0.1747384817 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 8 |
Hb_001532_060 |
0.1798227147 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003413_020 |
0.1837234899 |
- |
- |
- |
| 10 |
Hb_006262_030 |
0.1853765945 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_002805_260 |
0.1893063279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001288_020 |
0.1913590376 |
- |
- |
- |
| 13 |
Hb_150745_010 |
0.1950777447 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 14 |
Hb_008080_030 |
0.1998263334 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 15 |
Hb_006294_030 |
0.2034161935 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001159_220 |
0.2055796624 |
- |
- |
ESC, putative [Ricinus communis] |
| 17 |
Hb_002374_360 |
0.2091912291 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 18 |
Hb_000862_060 |
0.2120250149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_142734_020 |
0.2123114956 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_004316_160 |
0.2139070117 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |