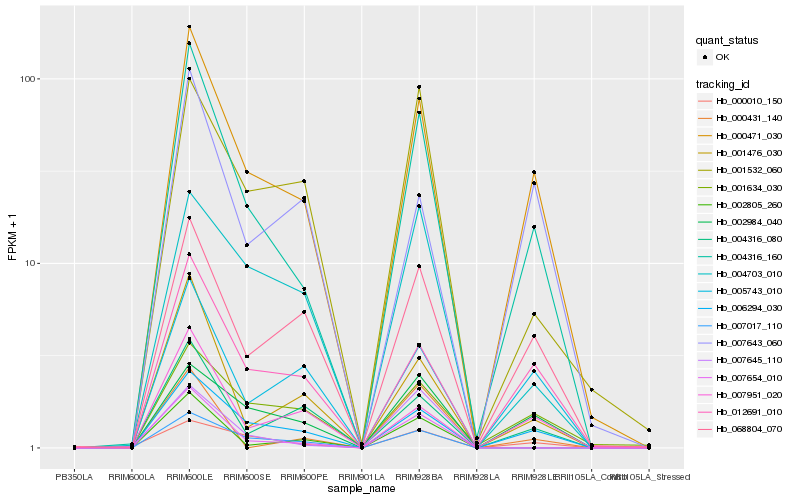
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004316_160 |
0.0 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 2 |
Hb_000471_030 |
0.1022420196 |
- |
- |
- |
| 3 |
Hb_006294_030 |
0.1362267335 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_012691_010 |
0.168119988 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 5 |
Hb_001476_030 |
0.1710899304 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_007654_010 |
0.1892379171 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 7 |
Hb_068804_070 |
0.1904487116 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 8 |
Hb_000010_150 |
0.1908282509 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 9 |
Hb_002984_040 |
0.2020066867 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_007951_020 |
0.2024818514 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 11 |
Hb_004316_080 |
0.2042653885 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 12 |
Hb_005743_010 |
0.2043619321 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 13 |
Hb_001634_030 |
0.2093870337 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 14 |
Hb_007017_110 |
0.2139070117 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 15 |
Hb_002805_260 |
0.2227768971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004703_010 |
0.22707352 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 17 |
Hb_000431_140 |
0.2281275026 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 18 |
Hb_007643_060 |
0.2313784327 |
- |
- |
- |
| 19 |
Hb_007645_110 |
0.2315037258 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 20 |
Hb_001532_060 |
0.2324412142 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |