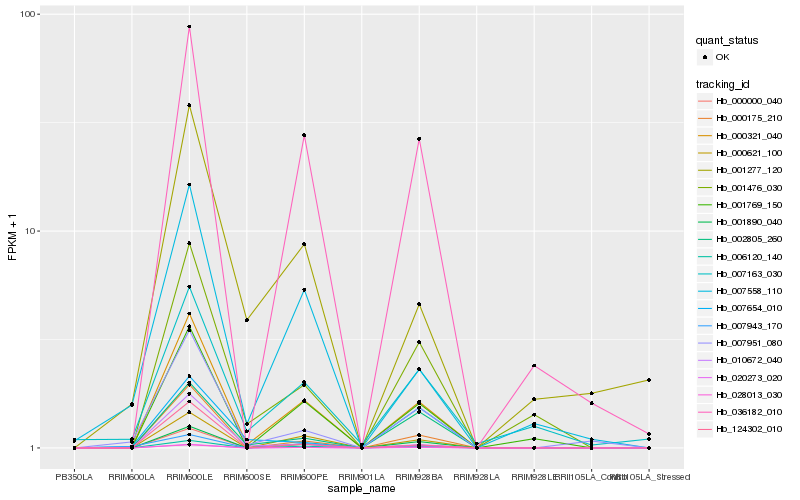
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007951_080 |
0.0 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 2 |
Hb_000175_210 |
0.1700940337 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 3 |
Hb_001890_040 |
0.1794520714 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_028013_030 |
0.1835737673 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_020273_020 |
0.186651924 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_000321_040 |
0.1867887711 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 7 |
Hb_002805_260 |
0.1993951415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006120_140 |
0.2077654015 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 9 |
Hb_007654_010 |
0.209705463 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 10 |
Hb_001476_030 |
0.2106564179 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_000000_040 |
0.2124958301 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 12 |
Hb_036182_010 |
0.232984942 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 13 |
Hb_001277_120 |
0.2362167156 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 14 |
Hb_010672_040 |
0.2362294694 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_124302_010 |
0.2369508246 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 16 |
Hb_000621_100 |
0.2371320308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001769_150 |
0.2389042064 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 18 |
Hb_007163_030 |
0.2406948671 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 19 |
Hb_007943_170 |
0.2407978002 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 20 |
Hb_007558_110 |
0.2409971324 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |