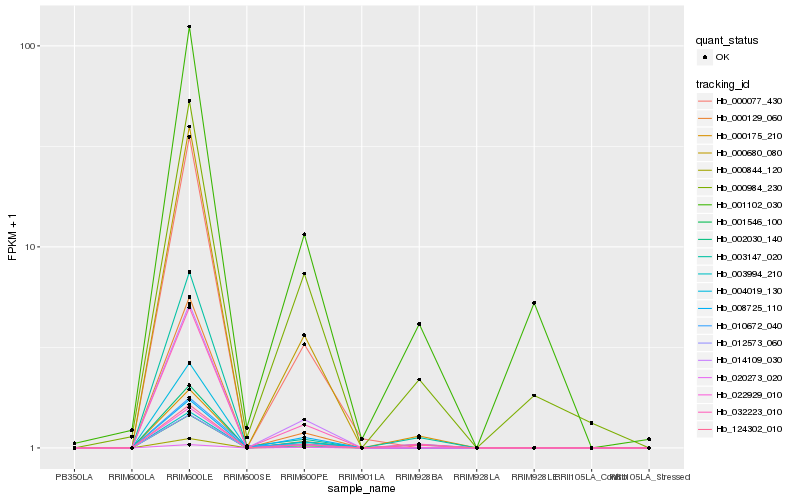
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124302_010 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 2 |
Hb_032223_010 |
0.0943968544 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 3 |
Hb_010672_040 |
0.1112097801 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_008725_110 |
0.1210828468 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_003147_020 |
0.1250027875 |
- |
- |
hypothetical protein POPTR_0019s07690g [Populus trichocarpa] |
| 6 |
Hb_000175_210 |
0.1250188972 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_000984_230 |
0.1433455577 |
- |
- |
- |
| 8 |
Hb_001102_030 |
0.1460568134 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 9 |
Hb_001546_100 |
0.1508359177 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 10 |
Hb_002030_140 |
0.1508759103 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 11 |
Hb_022929_010 |
0.1509207169 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 12 |
Hb_000680_080 |
0.150939024 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 13 |
Hb_012573_060 |
0.151141063 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 14 |
Hb_020273_020 |
0.1533681421 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_004019_130 |
0.15495591 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_014109_030 |
0.1555706699 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 17 |
Hb_000077_430 |
0.1556582411 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 18 |
Hb_000844_120 |
0.1562459462 |
- |
- |
hypothetical protein JCGZ_09179 [Jatropha curcas] |
| 19 |
Hb_000129_060 |
0.1563497333 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 20 |
Hb_003994_210 |
0.1563968618 |
- |
- |
zinc finger protein, putative [Ricinus communis] |