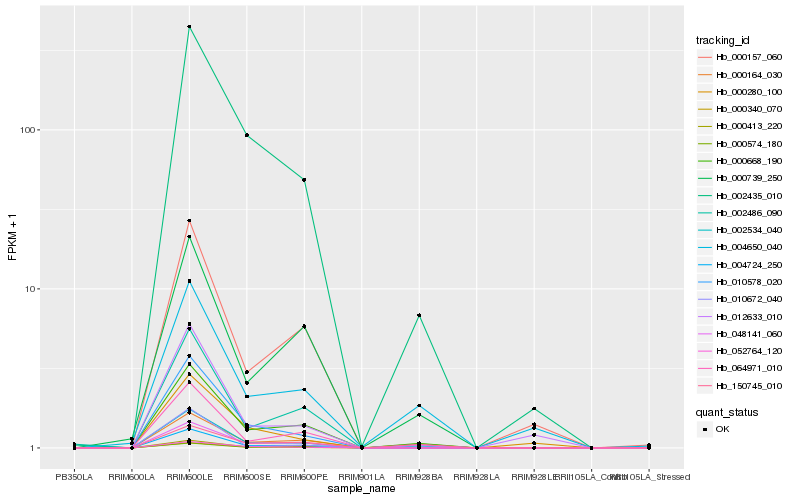
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_040 |
0.0 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 2 |
Hb_000668_190 |
0.0749439557 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 3 |
Hb_000340_070 |
0.1096468077 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 4 |
Hb_000413_220 |
0.1153109761 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 5 |
Hb_002435_010 |
0.1156104171 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 6 |
Hb_004724_250 |
0.1258250275 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_010672_040 |
0.1328754862 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_002486_090 |
0.1345128701 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 9 |
Hb_010578_020 |
0.1363177358 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000739_250 |
0.1395915333 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 11 |
Hb_048141_060 |
0.145639535 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 12 |
Hb_000164_030 |
0.1468759753 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 13 |
Hb_004650_040 |
0.1481503529 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 14 |
Hb_064971_010 |
0.1514121848 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 15 |
Hb_000280_100 |
0.1537363707 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 16 |
Hb_000574_180 |
0.1550000011 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 17 |
Hb_012633_010 |
0.1569514489 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 18 |
Hb_052764_120 |
0.1599159791 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 19 |
Hb_000157_060 |
0.1622926832 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_150745_010 |
0.1624511031 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |