| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
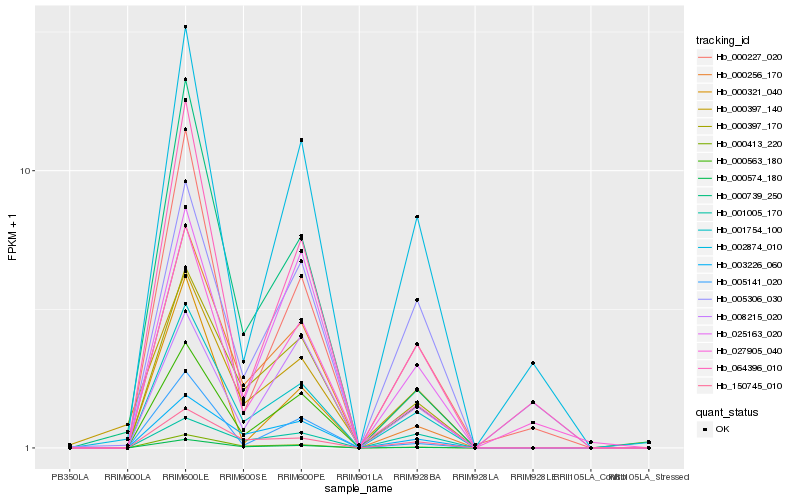
Hb_001754_100 |
0.0 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 2 |
Hb_000397_170 |
0.083309112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_150745_010 |
0.0906834626 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 4 |
Hb_003226_060 |
0.1024153102 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 5 |
Hb_000574_180 |
0.1026575839 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 6 |
Hb_000413_220 |
0.1133500595 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 7 |
Hb_000256_170 |
0.1341098076 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |
| 8 |
Hb_000227_020 |
0.1399375742 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 9 |
Hb_025163_020 |
0.1451789078 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 10 |
Hb_000563_180 |
0.1464345235 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 11 |
Hb_002874_010 |
0.1487771353 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000739_250 |
0.1537805833 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 13 |
Hb_064396_010 |
0.1539082001 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000397_140 |
0.1563146645 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 15 |
Hb_000321_040 |
0.1627337655 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 16 |
Hb_027905_040 |
0.1629873807 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 17 |
Hb_005141_020 |
0.1640039359 |
- |
- |
laccase, putative [Ricinus communis] |
| 18 |
Hb_005306_030 |
0.165041178 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 19 |
Hb_001005_170 |
0.1657813674 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 20 |
Hb_008215_020 |
0.1668008613 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |