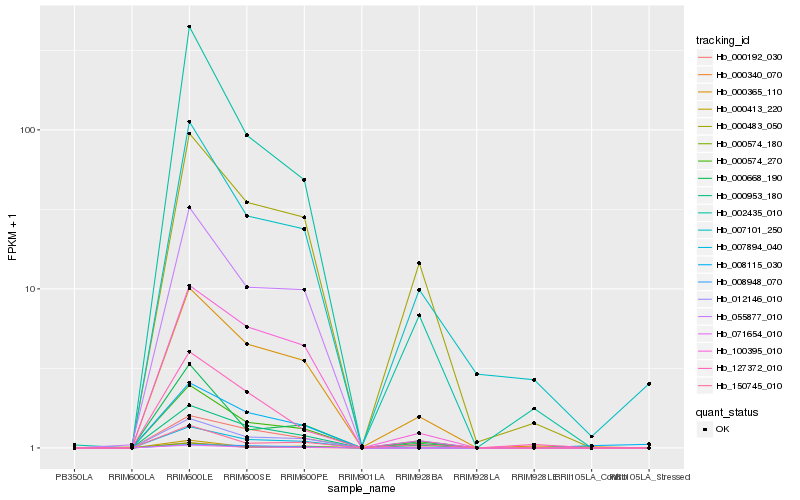
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_270 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 2 |
Hb_000365_110 |
0.0659134362 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 3 |
Hb_000953_180 |
0.0711156581 |
- |
- |
- |
| 4 |
Hb_000483_050 |
0.0966459725 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 5 |
Hb_150745_010 |
0.1150906573 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 6 |
Hb_000192_030 |
0.1196728763 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 7 |
Hb_000413_220 |
0.1236647384 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 8 |
Hb_100395_010 |
0.1259818309 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000340_070 |
0.1273712314 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 10 |
Hb_007101_250 |
0.1298069596 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 11 |
Hb_000574_180 |
0.1326946726 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 12 |
Hb_000668_190 |
0.1330279906 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 13 |
Hb_002435_010 |
0.1341827283 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 14 |
Hb_008115_030 |
0.1386513783 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_008948_070 |
0.1401048597 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_071654_010 |
0.142817333 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 17 |
Hb_012146_010 |
0.1455927863 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 18 |
Hb_127372_010 |
0.1471892451 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 19 |
Hb_007894_040 |
0.1481537763 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_055877_010 |
0.1492963186 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |