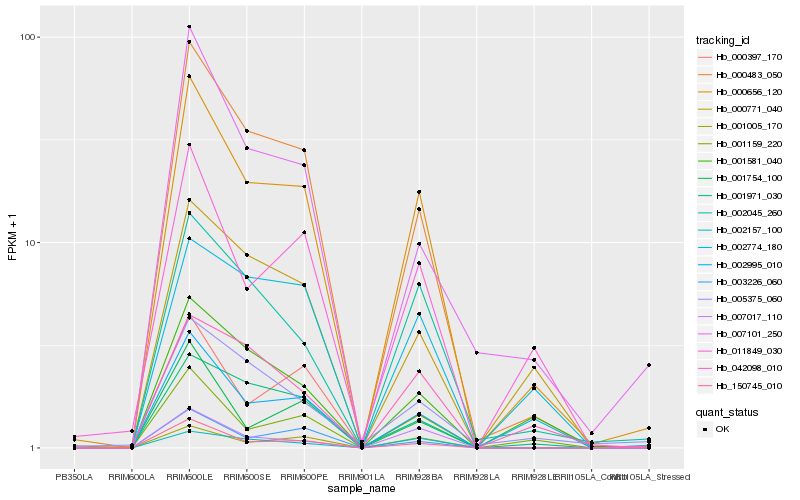
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_120 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 2 |
Hb_000483_050 |
0.1077542321 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 3 |
Hb_150745_010 |
0.1284319378 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 4 |
Hb_002045_260 |
0.1332828834 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 5 |
Hb_011849_030 |
0.138388488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001159_220 |
0.1384717556 |
- |
- |
ESC, putative [Ricinus communis] |
| 7 |
Hb_001005_170 |
0.143287006 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 8 |
Hb_001971_030 |
0.1444990419 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 9 |
Hb_001581_040 |
0.1488221872 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003226_060 |
0.1572969895 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 11 |
Hb_002157_100 |
0.1586083668 |
- |
- |
- |
| 12 |
Hb_007017_110 |
0.1588056056 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_000771_040 |
0.158820526 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 14 |
Hb_002774_180 |
0.1619509799 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_000397_170 |
0.164299018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007101_250 |
0.1646500773 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 17 |
Hb_042098_010 |
0.1652447038 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 18 |
Hb_001754_100 |
0.167623816 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 19 |
Hb_002995_010 |
0.1690967087 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 20 |
Hb_005375_060 |
0.1692044766 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |