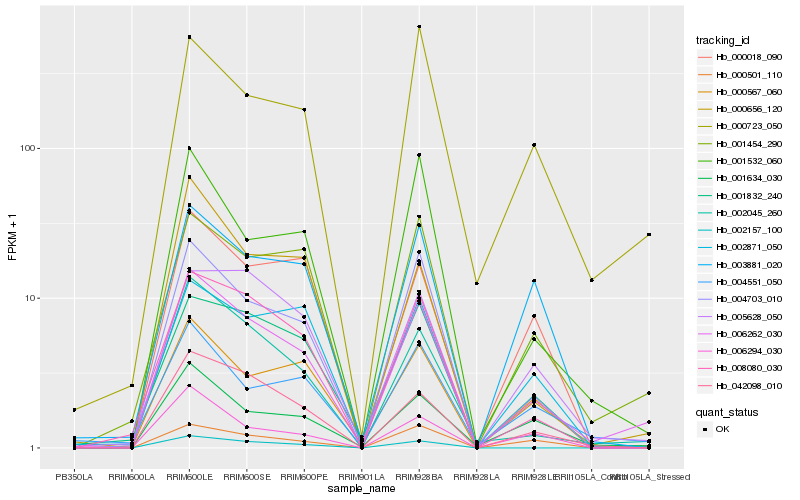
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006262_030 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_002045_260 |
0.119861647 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 3 |
Hb_004703_010 |
0.1284293299 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 4 |
Hb_042098_010 |
0.1359064381 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 5 |
Hb_008080_030 |
0.1401292742 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 6 |
Hb_001454_290 |
0.1441723179 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 7 |
Hb_001634_030 |
0.1467426611 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 8 |
Hb_001832_240 |
0.1579902499 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_000567_060 |
0.160071718 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 10 |
Hb_004551_050 |
0.1607838517 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002157_100 |
0.1612693346 |
- |
- |
- |
| 12 |
Hb_001532_060 |
0.1619273694 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000723_050 |
0.1628904811 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 14 |
Hb_005628_050 |
0.1666368989 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_006294_030 |
0.1667361305 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003881_020 |
0.1709816258 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 17 |
Hb_000501_110 |
0.1717676733 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 18 |
Hb_000656_120 |
0.1725900944 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 19 |
Hb_000018_090 |
0.1738763343 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_002871_050 |
0.1743274314 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |