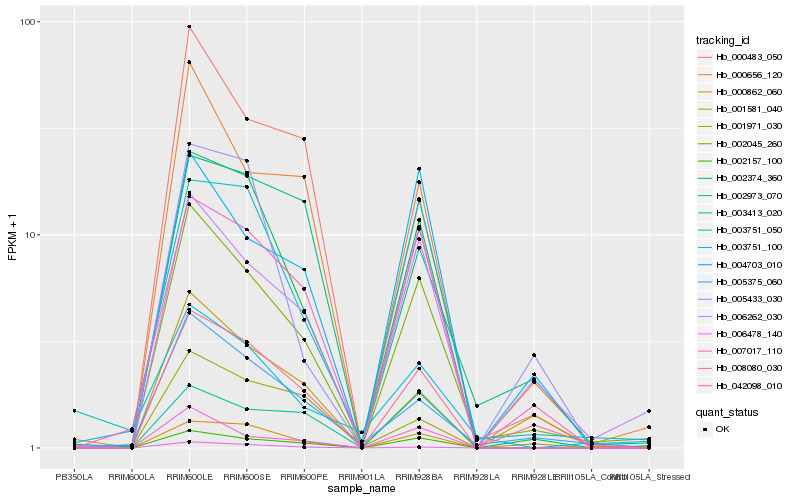
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_260 |
0.0 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 2 |
Hb_002157_100 |
0.114610547 |
- |
- |
- |
| 3 |
Hb_006262_030 |
0.119861647 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_042098_010 |
0.1244852084 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 5 |
Hb_000656_120 |
0.1332828834 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 6 |
Hb_002374_360 |
0.1361575528 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 7 |
Hb_007017_110 |
0.1406316974 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_008080_030 |
0.14143873 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 9 |
Hb_000862_060 |
0.1489410287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005375_060 |
0.151724424 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 11 |
Hb_004703_010 |
0.1673130164 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 12 |
Hb_001581_040 |
0.1686530946 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003751_050 |
0.1738324687 |
- |
- |
PREDICTED: amino acid permease 4-like [Jatropha curcas] |
| 14 |
Hb_000483_050 |
0.186426751 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 15 |
Hb_003413_020 |
0.1878730452 |
- |
- |
- |
| 16 |
Hb_005433_030 |
0.1880912983 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_006478_140 |
0.1932583986 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 18 |
Hb_001971_030 |
0.1940337971 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 19 |
Hb_003751_100 |
0.195544671 |
- |
- |
Amino acid permease 2 [Glycine soja] |
| 20 |
Hb_002973_070 |
0.1972286143 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |