| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
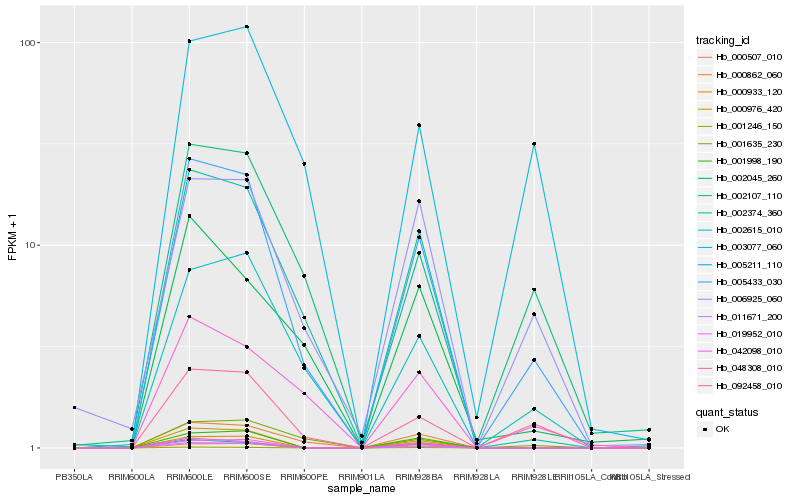
Hb_005433_030 |
0.0 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_092458_010 |
0.1229056562 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_002615_010 |
0.1285678361 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 4 |
Hb_002374_360 |
0.1332736334 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 5 |
Hb_002107_110 |
0.1566439679 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_042098_010 |
0.1593254272 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 7 |
Hb_006925_060 |
0.1595006892 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 8 |
Hb_000862_060 |
0.1643176747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000976_420 |
0.1693728132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001635_230 |
0.1702130016 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000933_120 |
0.1730196642 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 12 |
Hb_000507_010 |
0.1765759445 |
- |
- |
hypothetical protein POPTR_0007s02420g [Populus trichocarpa] |
| 13 |
Hb_019952_010 |
0.1783426687 |
- |
- |
- |
| 14 |
Hb_001246_150 |
0.1788921503 |
- |
- |
hypothetical protein MIMGU_mgv1a021562mg [Erythranthe guttata] |
| 15 |
Hb_005211_110 |
0.1813692254 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 16 |
Hb_048308_010 |
0.1817190996 |
- |
- |
Peroxidase superfamily protein [Theobroma cacao] |
| 17 |
Hb_003077_060 |
0.1832613827 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 18 |
Hb_011671_200 |
0.1834956938 |
- |
- |
- |
| 19 |
Hb_002045_260 |
0.1880912983 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 20 |
Hb_001998_190 |
0.1886543473 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |