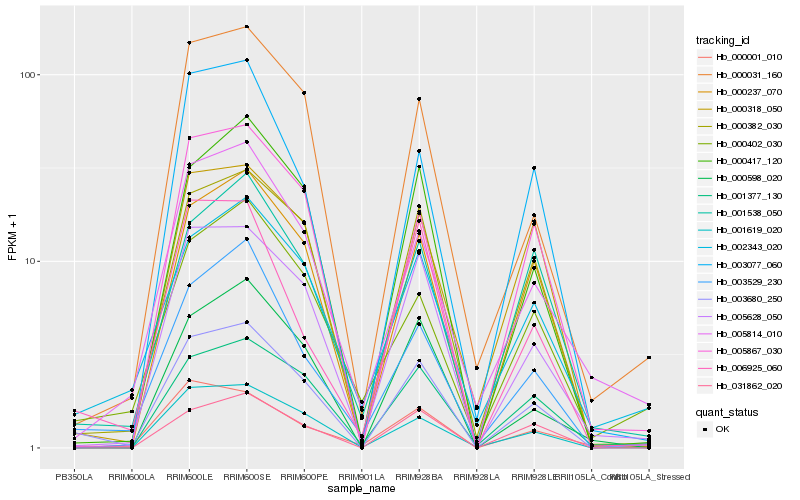
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_250 |
0.0 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 2 |
Hb_005628_050 |
0.1174716056 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_000382_030 |
0.1190009579 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 4 |
Hb_000237_070 |
0.1309784531 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 5 |
Hb_000417_120 |
0.1316535022 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 6 |
Hb_000031_160 |
0.1365097051 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 7 |
Hb_003529_230 |
0.1368999368 |
transcription factor |
TF Family: ERF |
- |
| 8 |
Hb_001377_130 |
0.1374100844 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 9 |
Hb_005867_030 |
0.1396294266 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_001619_020 |
0.1416958234 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 11 |
Hb_006925_060 |
0.147678237 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 12 |
Hb_031862_020 |
0.1477671983 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 13 |
Hb_002343_020 |
0.1501200093 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 14 |
Hb_000318_050 |
0.150649258 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001538_050 |
0.1523400241 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 16 |
Hb_000598_020 |
0.1544214 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 17 |
Hb_003077_060 |
0.154845669 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 18 |
Hb_000001_010 |
0.1596629804 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 19 |
Hb_005814_010 |
0.1599581071 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000402_030 |
0.160049777 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |