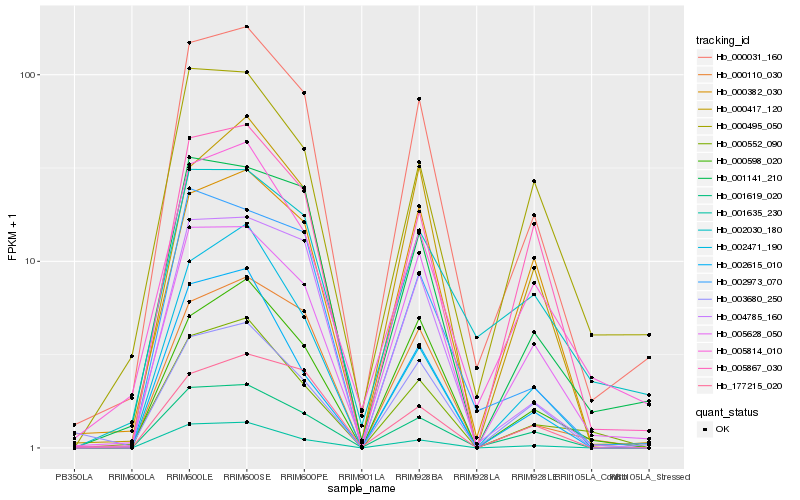
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_160 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 2 |
Hb_001619_020 |
0.0907850022 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 3 |
Hb_005628_050 |
0.1036569335 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_001141_210 |
0.10690173 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 5 |
Hb_005814_010 |
0.1116906564 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004785_160 |
0.1118485628 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 7 |
Hb_001635_230 |
0.1151385596 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000110_030 |
0.116076596 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002973_070 |
0.1239225706 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 10 |
Hb_000417_120 |
0.1241404377 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 11 |
Hb_000598_020 |
0.1248157067 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 12 |
Hb_000552_090 |
0.1256970497 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 13 |
Hb_005867_030 |
0.1285631254 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_002030_180 |
0.1338571343 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_000495_050 |
0.1343925226 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 16 |
Hb_003680_250 |
0.1365097051 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 17 |
Hb_002615_010 |
0.140701965 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 18 |
Hb_002471_190 |
0.141315 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 19 |
Hb_177215_020 |
0.1415002902 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 20 |
Hb_000382_030 |
0.1433310114 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |