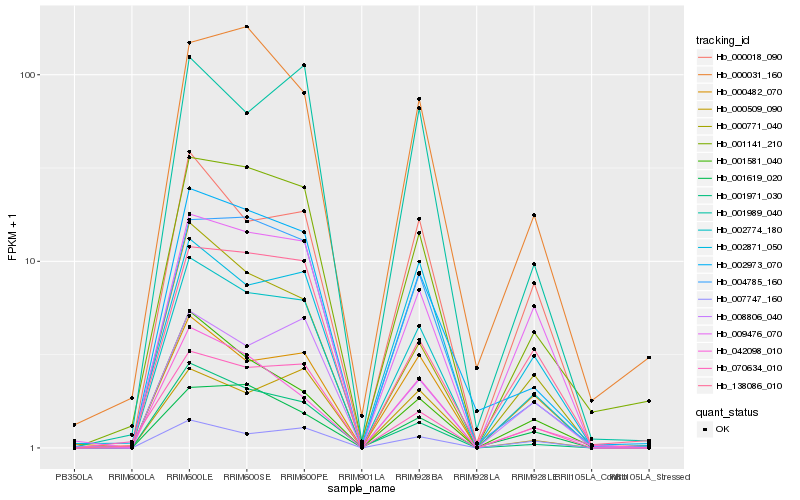
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_180 |
0.0 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_002973_070 |
0.0995307963 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 3 |
Hb_000018_090 |
0.1031437364 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001141_210 |
0.1035549976 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 5 |
Hb_000771_040 |
0.1093431379 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 6 |
Hb_009476_070 |
0.1121116574 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 7 |
Hb_004785_160 |
0.1154005724 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 8 |
Hb_008806_040 |
0.1157038931 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 9 |
Hb_001971_030 |
0.1228707561 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 10 |
Hb_001989_040 |
0.1238353542 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 11 |
Hb_000482_070 |
0.1238900521 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 12 |
Hb_070634_010 |
0.1252657299 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 13 |
Hb_138086_010 |
0.1323319748 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_042098_010 |
0.1325049102 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 15 |
Hb_001619_020 |
0.1329557376 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 16 |
Hb_002871_050 |
0.1340137327 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 17 |
Hb_000509_090 |
0.1369132403 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 18 |
Hb_007747_160 |
0.1439895867 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_001581_040 |
0.1440137791 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000031_160 |
0.1454648264 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |