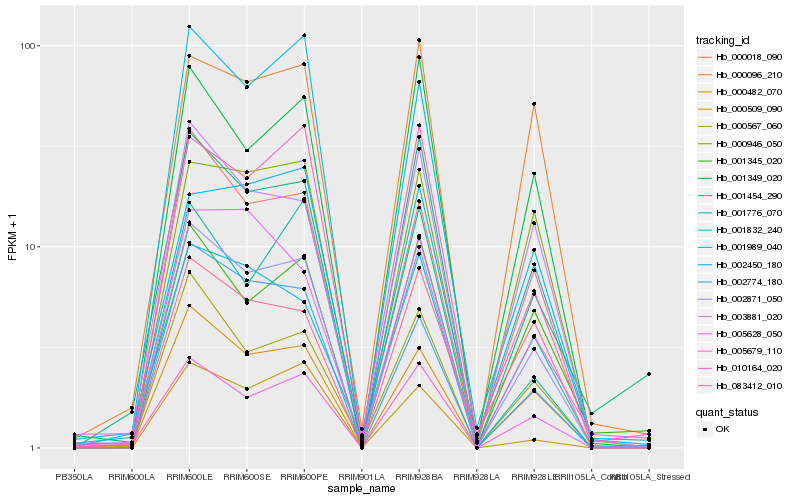
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002871_050 |
0.0 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 2 |
Hb_000482_070 |
0.0775689304 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 3 |
Hb_005679_110 |
0.0781527513 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 4 |
Hb_000567_060 |
0.0949102877 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 5 |
Hb_000018_090 |
0.0980454811 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001832_240 |
0.1064182971 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 7 |
Hb_003881_020 |
0.1080382674 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 8 |
Hb_001454_290 |
0.1092899548 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 9 |
Hb_010164_020 |
0.112994867 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 10 |
Hb_001349_020 |
0.1154708199 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 11 |
Hb_001776_070 |
0.1220092358 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 12 |
Hb_001345_020 |
0.1229439607 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 13 |
Hb_001989_040 |
0.127371784 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 14 |
Hb_002774_180 |
0.1340137327 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_000096_210 |
0.135010519 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 16 |
Hb_000509_090 |
0.1380042258 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_002450_180 |
0.1407959881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000946_050 |
0.1409254147 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_083412_010 |
0.1413431475 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_005628_050 |
0.1446001117 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |