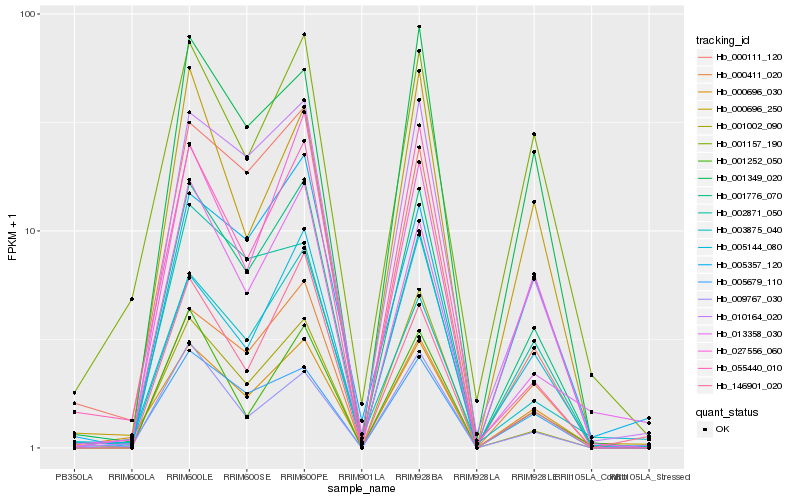
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_070 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 2 |
Hb_010164_020 |
0.085102893 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 3 |
Hb_000696_030 |
0.0944920491 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 4 |
Hb_027556_060 |
0.0980323796 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 5 |
Hb_005679_110 |
0.0994324576 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 6 |
Hb_001349_020 |
0.1126683632 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 7 |
Hb_005144_080 |
0.1157300084 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_001252_050 |
0.1177795413 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 9 |
Hb_002871_050 |
0.1220092358 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 10 |
Hb_013358_030 |
0.12230004 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 11 |
Hb_003875_040 |
0.1228791531 |
- |
- |
PREDICTED: endoglucanase 11-like [Jatropha curcas] |
| 12 |
Hb_000111_120 |
0.1240935774 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001002_090 |
0.1259543177 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 14 |
Hb_055440_010 |
0.1263769949 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000696_250 |
0.1278934138 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 16 |
Hb_009767_030 |
0.1289453352 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 17 |
Hb_001157_190 |
0.1294809114 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 18 |
Hb_000411_020 |
0.1304521196 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_146901_020 |
0.1319440843 |
- |
- |
- |
| 20 |
Hb_005357_120 |
0.1342240725 |
transcription factor |
TF Family: MYB-related |
- |