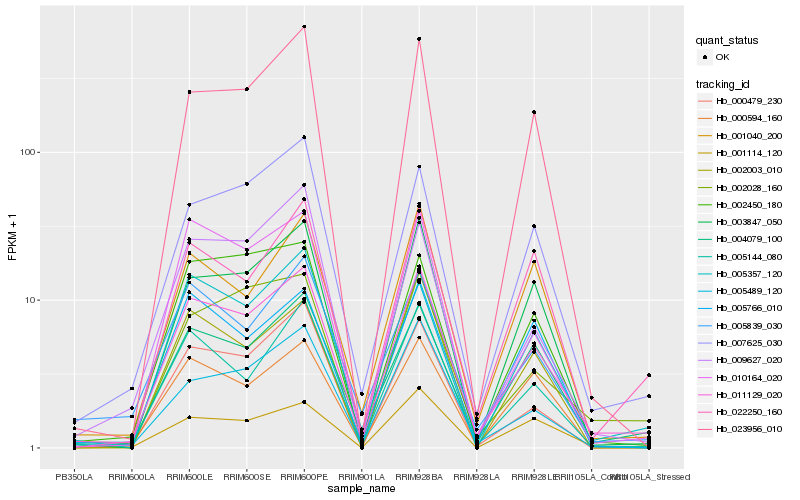
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011129_020 |
0.0 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 2 |
Hb_004079_100 |
0.0987369142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 3 |
Hb_023956_010 |
0.1110744687 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 4 |
Hb_005144_080 |
0.111111296 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 5 |
Hb_003847_050 |
0.1119895103 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 6 |
Hb_000479_230 |
0.1139282206 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 7 |
Hb_007625_030 |
0.1146477239 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_010164_020 |
0.1157540757 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_022250_160 |
0.1220279973 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 10 |
Hb_000594_160 |
0.1231699239 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 11 |
Hb_005839_030 |
0.1257214125 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 12 |
Hb_005357_120 |
0.1268420149 |
transcription factor |
TF Family: MYB-related |
- |
| 13 |
Hb_001040_200 |
0.1276519374 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 14 |
Hb_005489_120 |
0.1298767312 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 15 |
Hb_002028_160 |
0.1312113356 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |
| 16 |
Hb_001114_120 |
0.1340445932 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002450_180 |
0.1342188396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005766_010 |
0.1356315624 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_009627_020 |
0.1377419596 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 20 |
Hb_002003_010 |
0.1378106784 |
- |
- |
hypothetical protein JCGZ_15062 [Jatropha curcas] |