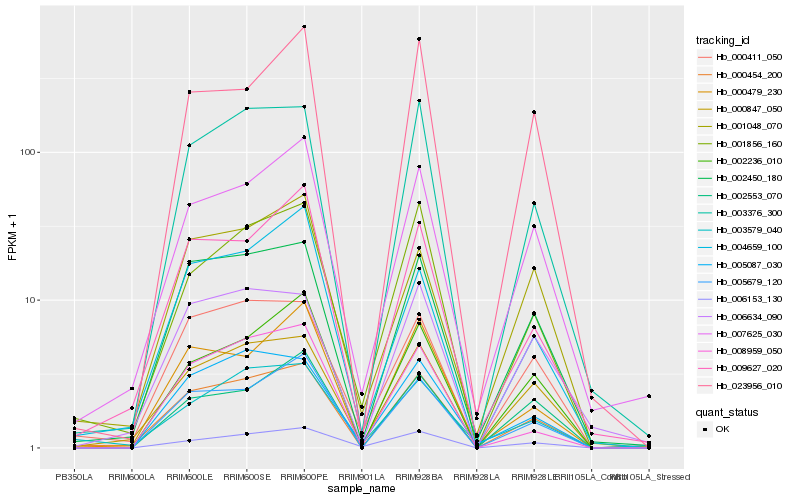
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000454_200 |
0.0 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 2 |
Hb_002450_180 |
0.0807292325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000847_050 |
0.0849525452 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 4 |
Hb_003376_300 |
0.0903343554 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 5 |
Hb_005679_120 |
0.1003931522 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 6 |
Hb_000411_050 |
0.1078322102 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 7 |
Hb_001856_160 |
0.1123501157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007625_030 |
0.1128798176 |
- |
- |
unknown [Medicago truncatula] |
| 9 |
Hb_009627_020 |
0.1166264041 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 10 |
Hb_002236_010 |
0.1242096677 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 11 |
Hb_000479_230 |
0.1248805481 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 12 |
Hb_006634_090 |
0.1299560887 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 13 |
Hb_005087_030 |
0.1306410532 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_001048_070 |
0.1314897309 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 15 |
Hb_023956_010 |
0.1318202661 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_004659_100 |
0.1321138641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006153_130 |
0.1325446138 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_008959_050 |
0.1344998317 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002553_070 |
0.1356072608 |
- |
- |
PREDICTED: RING-H2 finger protein ATL16 [Jatropha curcas] |
| 20 |
Hb_003579_040 |
0.1357205857 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |