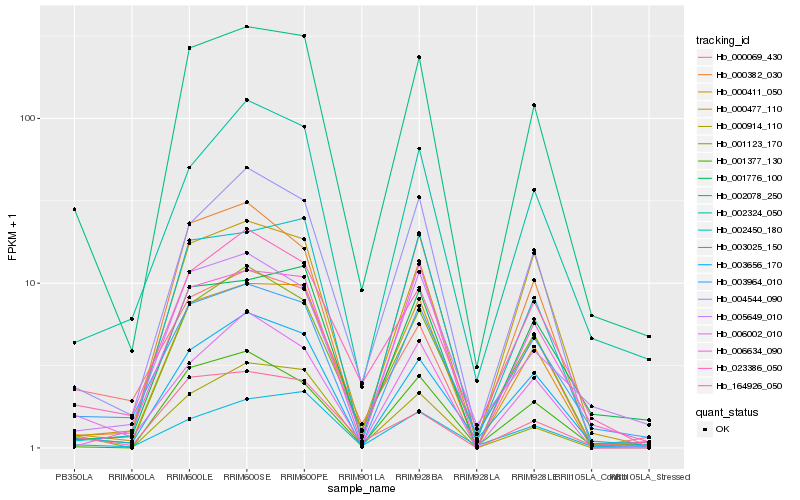
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000411_050 |
0.084698556 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 3 |
Hb_003656_170 |
0.0991381077 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 4 |
Hb_003964_010 |
0.0998427642 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 5 |
Hb_001123_170 |
0.1036093624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004544_090 |
0.1119795078 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_003025_150 |
0.1133534198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001776_100 |
0.120003771 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002450_180 |
0.1203447569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002324_050 |
0.1224989455 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 11 |
Hb_023386_050 |
0.1235556022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000914_110 |
0.1237307113 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 13 |
Hb_001377_130 |
0.1247644329 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 14 |
Hb_000069_430 |
0.1260270763 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 15 |
Hb_164926_050 |
0.1283645722 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 16 |
Hb_000382_030 |
0.1322299683 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 17 |
Hb_000477_110 |
0.1334498844 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 18 |
Hb_005649_010 |
0.1354382908 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_006002_010 |
0.138420995 |
transcription factor |
TF Family: MYB |
Myb domain protein 5 isoform 1 [Theobroma cacao] |
| 20 |
Hb_006634_090 |
0.13975511 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |