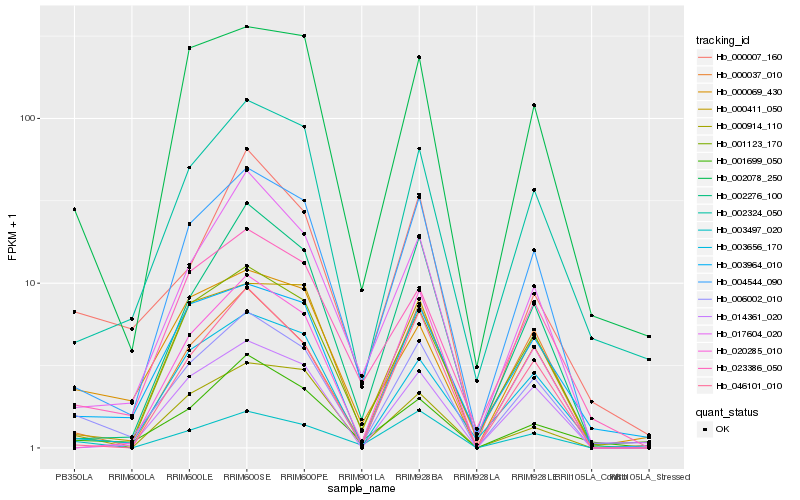
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006002_010 |
0.0 |
transcription factor |
TF Family: MYB |
Myb domain protein 5 isoform 1 [Theobroma cacao] |
| 2 |
Hb_004544_090 |
0.0996285499 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_003497_020 |
0.1298640321 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 4 |
Hb_002078_250 |
0.138420995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_017604_020 |
0.1389742079 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 6 |
Hb_002324_050 |
0.1403826476 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 7 |
Hb_000069_430 |
0.1421191201 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 8 |
Hb_003964_010 |
0.1451949846 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 9 |
Hb_002276_100 |
0.1463615034 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 10 |
Hb_020285_010 |
0.1469876009 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 11 |
Hb_001699_050 |
0.1477784323 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 12 |
Hb_001123_170 |
0.1489328034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_046101_010 |
0.1492906633 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 14 |
Hb_000914_110 |
0.1506470773 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 15 |
Hb_000411_050 |
0.1510457638 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 16 |
Hb_003656_170 |
0.1510961684 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 17 |
Hb_000037_010 |
0.1511950463 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 18 |
Hb_014361_020 |
0.151811334 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 19 |
Hb_000007_160 |
0.1542730136 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 20 |
Hb_023386_050 |
0.1554652932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |