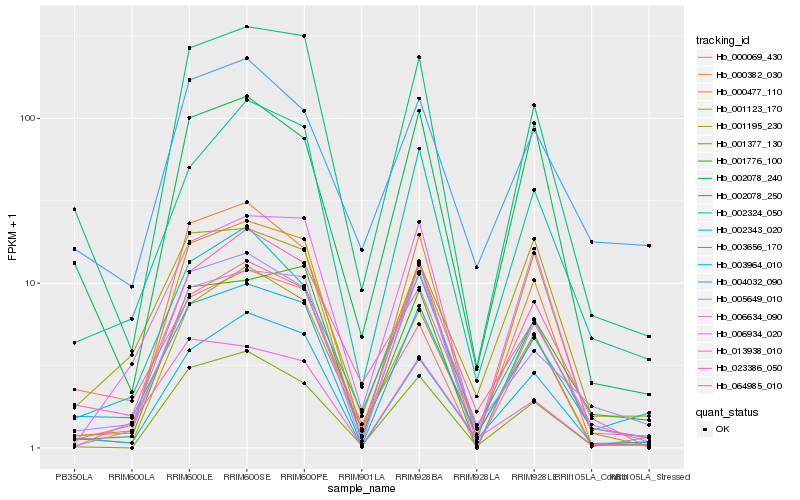
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_010 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 2 |
Hb_002078_250 |
0.0998427642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003656_170 |
0.1016373961 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 4 |
Hb_002324_050 |
0.1045629657 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_005649_010 |
0.1117554009 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 6 |
Hb_064985_010 |
0.1142279868 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 7 |
Hb_002343_020 |
0.1210162442 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 8 |
Hb_001123_170 |
0.1239224916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006934_020 |
0.1309026503 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 10 |
Hb_006634_090 |
0.1314469866 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 11 |
Hb_001776_100 |
0.1319158909 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002078_240 |
0.132075832 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 13 |
Hb_013938_010 |
0.1328283757 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 14 |
Hb_023386_050 |
0.1345793369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001377_130 |
0.1362199495 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 16 |
Hb_000069_430 |
0.1363215114 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 17 |
Hb_004032_090 |
0.1367970413 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 18 |
Hb_000382_030 |
0.1374581906 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 19 |
Hb_001195_230 |
0.1380102987 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 20 |
Hb_000477_110 |
0.1383963892 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |