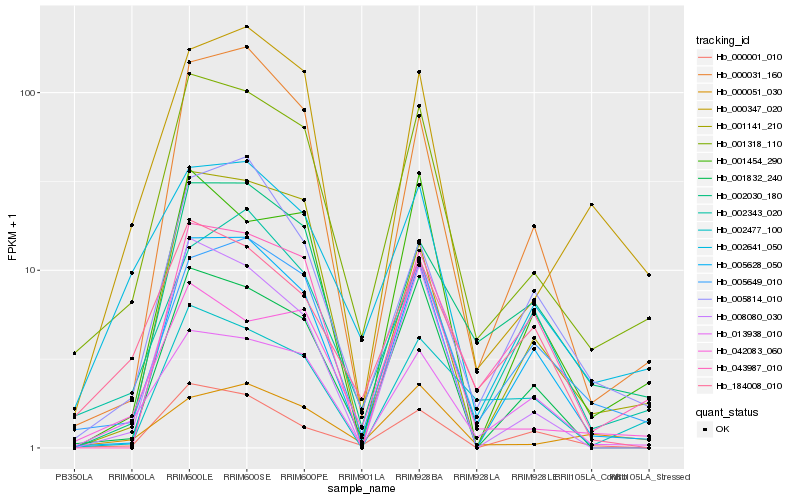
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 2 |
Hb_005649_010 |
0.1308053885 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 3 |
Hb_002641_050 |
0.1372406527 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 4 |
Hb_013938_010 |
0.1391318779 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 5 |
Hb_001832_240 |
0.1461256083 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 6 |
Hb_005628_050 |
0.1469582324 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_000031_160 |
0.1510361544 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 8 |
Hb_000347_020 |
0.1517682881 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 9 |
Hb_002030_180 |
0.1555946395 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_008080_030 |
0.1601785274 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 11 |
Hb_001141_210 |
0.160711639 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 12 |
Hb_184008_010 |
0.1633887185 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 13 |
Hb_043987_010 |
0.1689112216 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 14 |
Hb_000051_030 |
0.1699038212 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 15 |
Hb_000001_010 |
0.1704900055 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 16 |
Hb_001454_290 |
0.1724064101 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 17 |
Hb_002343_020 |
0.1733085054 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 18 |
Hb_005814_010 |
0.1735478288 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_042083_060 |
0.1742900408 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 20 |
Hb_002477_100 |
0.1764850738 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |