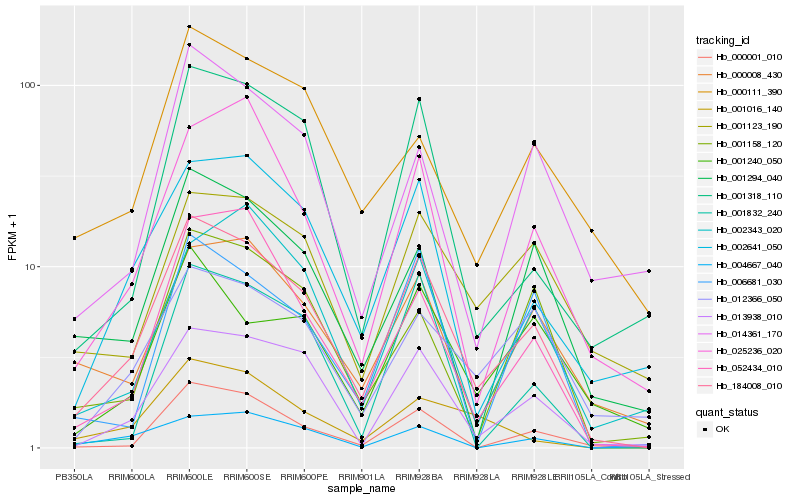
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184008_010 |
0.0 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 2 |
Hb_013938_010 |
0.129386055 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 3 |
Hb_002641_050 |
0.155324341 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 4 |
Hb_000001_010 |
0.1594857058 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 5 |
Hb_052434_010 |
0.1610876856 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 6 |
Hb_001318_110 |
0.1633887185 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 7 |
Hb_000008_430 |
0.1697181762 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 8 |
Hb_025236_020 |
0.1698183351 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 9 |
Hb_001294_040 |
0.1717912102 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 10 |
Hb_012366_050 |
0.1766422338 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 11 |
Hb_001158_120 |
0.1790218603 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001240_050 |
0.1810316211 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 13 |
Hb_001123_190 |
0.182550028 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 14 |
Hb_006681_030 |
0.1827516325 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000111_390 |
0.1846998686 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002343_020 |
0.1850583672 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 17 |
Hb_001016_140 |
0.1851450854 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 18 |
Hb_004667_040 |
0.185837991 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_014361_170 |
0.1872457222 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 20 |
Hb_001832_240 |
0.1894231166 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |