| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
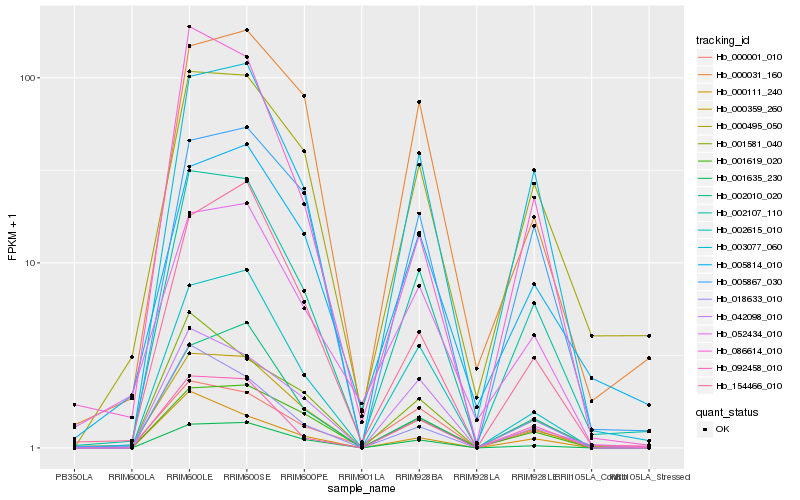
Hb_002107_110 |
0.0 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003077_060 |
0.0887337735 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 3 |
Hb_000359_260 |
0.0978678371 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 4 |
Hb_092458_010 |
0.1064330574 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 5 |
Hb_000495_050 |
0.1083867329 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 6 |
Hb_001635_230 |
0.11415404 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002615_010 |
0.1187556079 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 8 |
Hb_001619_020 |
0.1293545055 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 9 |
Hb_005867_030 |
0.1310684124 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_002010_020 |
0.1342971246 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 11 |
Hb_042098_010 |
0.1347364939 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_005814_010 |
0.1355690163 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_018633_010 |
0.1374029466 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001581_040 |
0.1385982097 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_052434_010 |
0.1397790948 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 16 |
Hb_000001_010 |
0.1408915842 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 17 |
Hb_000111_240 |
0.1419002043 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 18 |
Hb_154466_010 |
0.1445955261 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000031_160 |
0.1481654875 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 20 |
Hb_086614_010 |
0.1496041688 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |