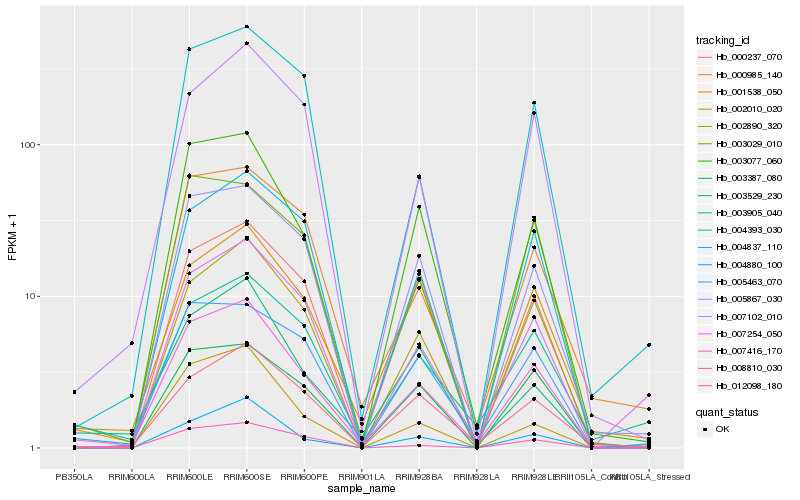
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_170 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 2 |
Hb_003077_060 |
0.1110501066 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 3 |
Hb_002890_320 |
0.1175752193 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 4 |
Hb_004880_100 |
0.1286333076 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 5 |
Hb_005867_030 |
0.1310017751 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 6 |
Hb_004837_110 |
0.131143422 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003029_010 |
0.1336055006 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007102_010 |
0.1336279108 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 9 |
Hb_000985_140 |
0.1353059093 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 10 |
Hb_012098_180 |
0.1361942843 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 11 |
Hb_003905_040 |
0.1369579108 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 12 |
Hb_003529_230 |
0.1382882679 |
transcription factor |
TF Family: ERF |
- |
| 13 |
Hb_001538_050 |
0.1396634033 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 14 |
Hb_008810_030 |
0.1404221318 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_003387_080 |
0.1423244049 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_004393_030 |
0.143929556 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 17 |
Hb_000237_070 |
0.144513543 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 18 |
Hb_002010_020 |
0.1447591958 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 19 |
Hb_005463_070 |
0.1459078358 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_007254_050 |
0.1467412251 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |