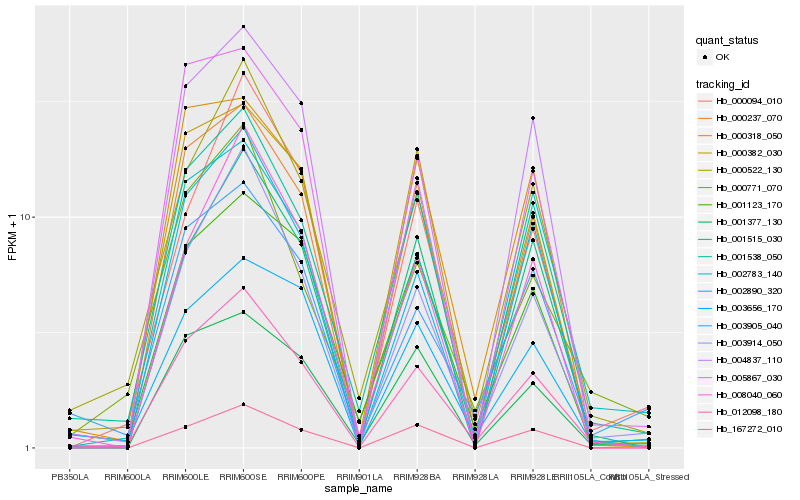
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_180 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 2 |
Hb_001538_050 |
0.0906766585 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 3 |
Hb_000237_070 |
0.0973511331 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 4 |
Hb_000522_130 |
0.1002295685 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 5 |
Hb_003914_050 |
0.1002732828 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 6 |
Hb_002890_320 |
0.1089721903 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 7 |
Hb_001123_170 |
0.1139341343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001515_030 |
0.1141346627 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001377_130 |
0.118020197 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 10 |
Hb_004837_110 |
0.1189812175 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_167272_010 |
0.120386382 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 12 |
Hb_000382_030 |
0.121058487 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 13 |
Hb_002783_140 |
0.1217925326 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 14 |
Hb_003905_040 |
0.1219600628 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 15 |
Hb_005867_030 |
0.1223453919 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 16 |
Hb_003656_170 |
0.1258277345 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 17 |
Hb_000318_050 |
0.1265666899 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000094_010 |
0.1274886947 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_008040_060 |
0.131078374 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 20 |
Hb_000771_070 |
0.1335887535 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |