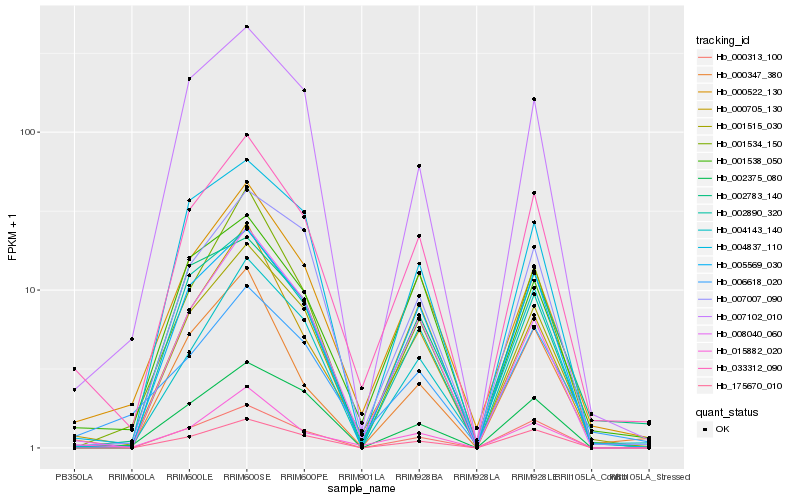
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_090 |
0.0 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000522_130 |
0.0929385123 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 3 |
Hb_015882_020 |
0.1015911268 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_005569_030 |
0.1079776267 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000705_130 |
0.1157681544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001538_050 |
0.1170452021 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 7 |
Hb_008040_060 |
0.1200517697 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_000313_100 |
0.121329936 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 9 |
Hb_002890_320 |
0.1218862874 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 10 |
Hb_006618_020 |
0.1236476481 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 11 |
Hb_175670_010 |
0.1240639341 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_002375_080 |
0.1261239308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007007_090 |
0.126861264 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 14 |
Hb_001515_030 |
0.1269269762 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004837_110 |
0.132003464 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007102_010 |
0.1322412647 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 17 |
Hb_001534_150 |
0.1344813176 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 18 |
Hb_002783_140 |
0.1362752201 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 19 |
Hb_004143_140 |
0.1369724427 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 20 |
Hb_000347_380 |
0.1383349392 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |