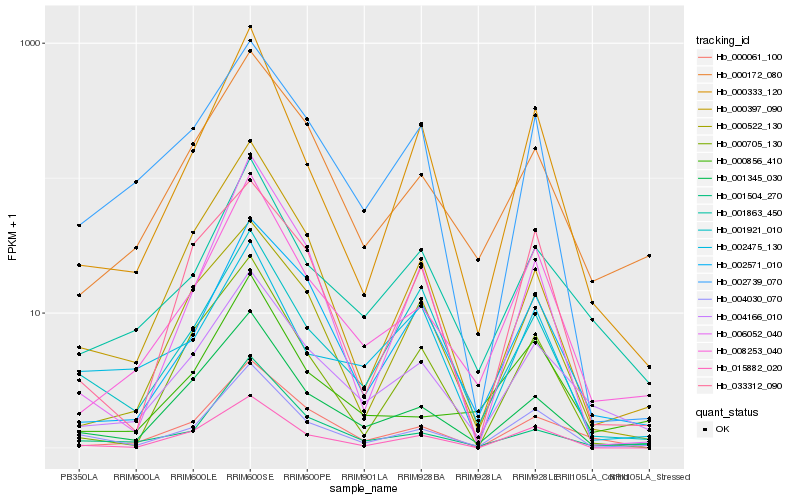
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004030_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 2 |
Hb_008253_040 |
0.1316328468 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 3 |
Hb_015882_020 |
0.1321954165 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_000333_120 |
0.1430563669 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 5 |
Hb_001345_030 |
0.1434081772 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 6 |
Hb_004166_010 |
0.1540456047 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 7 |
Hb_001504_270 |
0.1612865355 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 8 |
Hb_001921_010 |
0.1646220841 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000856_410 |
0.166703385 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_002571_010 |
0.1668021842 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000172_080 |
0.1692037021 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 12 |
Hb_001863_450 |
0.1695932773 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 13 |
Hb_002739_070 |
0.1711867815 |
- |
- |
hypothetical protein B456_002G169900 [Gossypium raimondii] |
| 14 |
Hb_000397_090 |
0.172734745 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 15 |
Hb_006052_040 |
0.1740479161 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 16 |
Hb_033312_090 |
0.1755607622 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002475_130 |
0.1810650149 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 18 |
Hb_000705_130 |
0.1844440603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000061_100 |
0.1872831681 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 20 |
Hb_000522_130 |
0.1884163663 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |