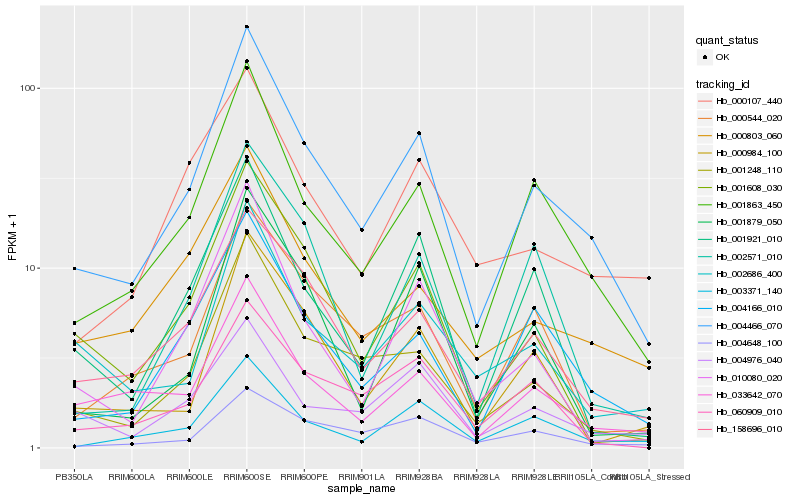
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004466_070 |
0.0 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 2 |
Hb_001863_450 |
0.0913087089 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 3 |
Hb_158696_010 |
0.1247616308 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 4 |
Hb_001248_110 |
0.1255627544 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_004166_010 |
0.1263653585 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 6 |
Hb_033642_070 |
0.1392376997 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 7 |
Hb_003371_140 |
0.1444752198 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 8 |
Hb_000803_060 |
0.1472392318 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 9 |
Hb_000544_020 |
0.1493116717 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 10 |
Hb_001608_030 |
0.1500128251 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 11 |
Hb_001921_010 |
0.153087168 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_010080_020 |
0.1554410798 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 13 |
Hb_002686_400 |
0.1569866153 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 14 |
Hb_004648_100 |
0.1582834823 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 15 |
Hb_000107_440 |
0.1593175679 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_000984_100 |
0.1593959351 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_004976_040 |
0.1598264958 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 18 |
Hb_001879_050 |
0.159984839 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 19 |
Hb_060909_010 |
0.1623540371 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 20 |
Hb_002571_010 |
0.1628737272 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |