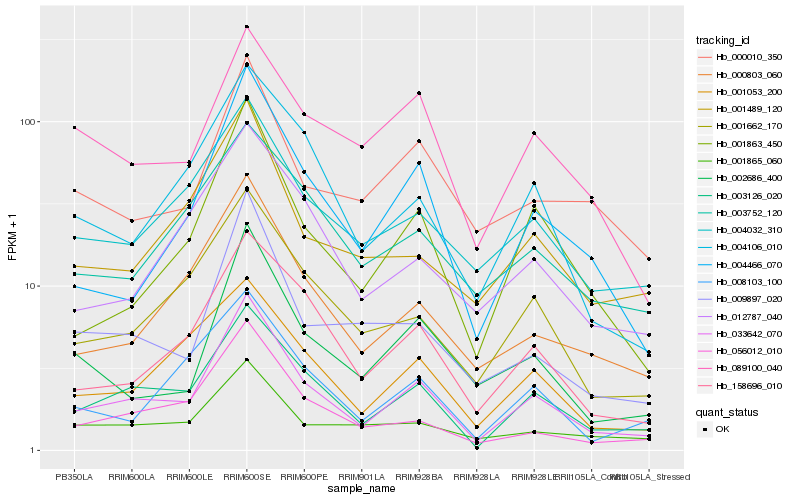
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033642_070 |
0.0 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 2 |
Hb_003126_020 |
0.0898954648 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 3 |
Hb_158696_010 |
0.1349318086 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 4 |
Hb_001863_450 |
0.1377852516 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 5 |
Hb_001662_170 |
0.1380578516 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 6 |
Hb_056012_010 |
0.1391114535 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 7 |
Hb_004466_070 |
0.1392376997 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 8 |
Hb_000803_060 |
0.1400491847 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 9 |
Hb_002686_400 |
0.1453625343 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 10 |
Hb_004106_010 |
0.1470513534 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009897_020 |
0.14844489 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_004032_310 |
0.1486034032 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 13 |
Hb_001053_200 |
0.1494034562 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 14 |
Hb_008103_100 |
0.1567025096 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 15 |
Hb_001489_120 |
0.1589377448 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_089100_040 |
0.1606823724 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 17 |
Hb_012787_040 |
0.1610811441 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001865_060 |
0.1629260707 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 19 |
Hb_000010_350 |
0.1653720139 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 20 |
Hb_003752_120 |
0.1680757705 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |