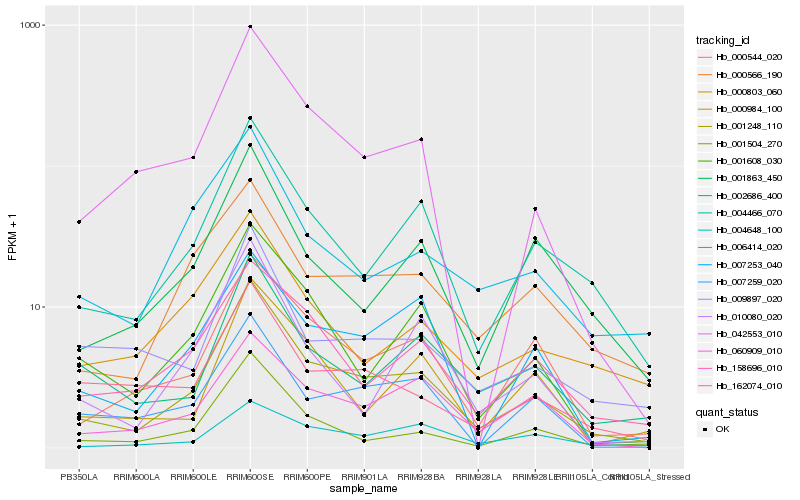
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001248_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_004466_070 |
0.1255627544 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 3 |
Hb_000544_020 |
0.1383865155 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_000566_190 |
0.1521015299 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001608_030 |
0.152666011 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 6 |
Hb_042553_010 |
0.1531139023 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 7 |
Hb_158696_010 |
0.1555986985 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 8 |
Hb_001504_270 |
0.1570284405 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 9 |
Hb_060909_010 |
0.161124222 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 10 |
Hb_002686_400 |
0.1634352438 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 11 |
Hb_001863_450 |
0.1639415583 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 12 |
Hb_006414_020 |
0.1642546471 |
transcription factor |
TF Family: MYB |
- |
| 13 |
Hb_007259_020 |
0.1697378848 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 14 |
Hb_009897_020 |
0.1733796795 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_162074_010 |
0.1737000914 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_010080_020 |
0.1752239514 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 17 |
Hb_007253_040 |
0.1752776442 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000984_100 |
0.1761657637 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_004648_100 |
0.1764953047 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 20 |
Hb_000803_060 |
0.1780304487 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |