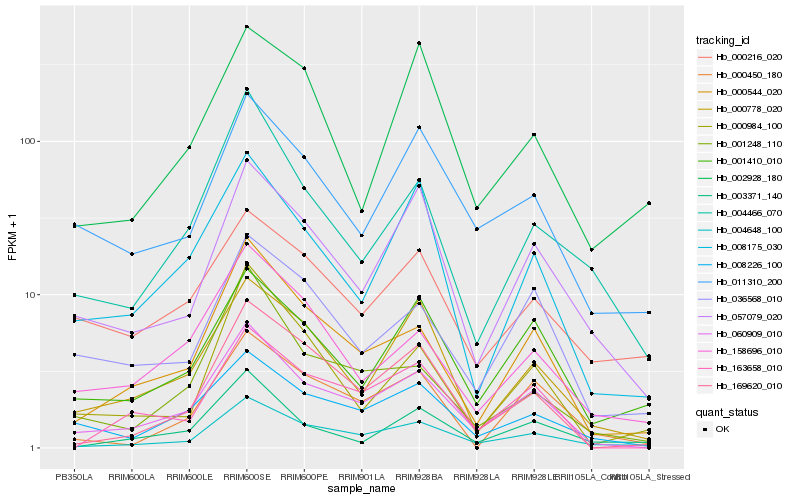
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004648_100 |
0.0 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 2 |
Hb_060909_010 |
0.1243360874 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 3 |
Hb_000544_020 |
0.1316537726 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_169620_010 |
0.1342488126 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 5 |
Hb_057079_020 |
0.1357786748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001410_010 |
0.1472022277 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 7 |
Hb_011310_200 |
0.1521978341 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 8 |
Hb_000450_180 |
0.153360291 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_008226_100 |
0.1555171033 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 10 |
Hb_000778_020 |
0.1566926542 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 11 |
Hb_004466_070 |
0.1582834823 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 12 |
Hb_003371_140 |
0.1652081147 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 13 |
Hb_002928_180 |
0.1659011934 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 14 |
Hb_158696_010 |
0.1671675327 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 15 |
Hb_163658_010 |
0.1675037593 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_036568_010 |
0.170507577 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 17 |
Hb_000216_020 |
0.1713174784 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 18 |
Hb_000984_100 |
0.175952807 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_001248_110 |
0.1764953047 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_008175_030 |
0.1770381382 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |