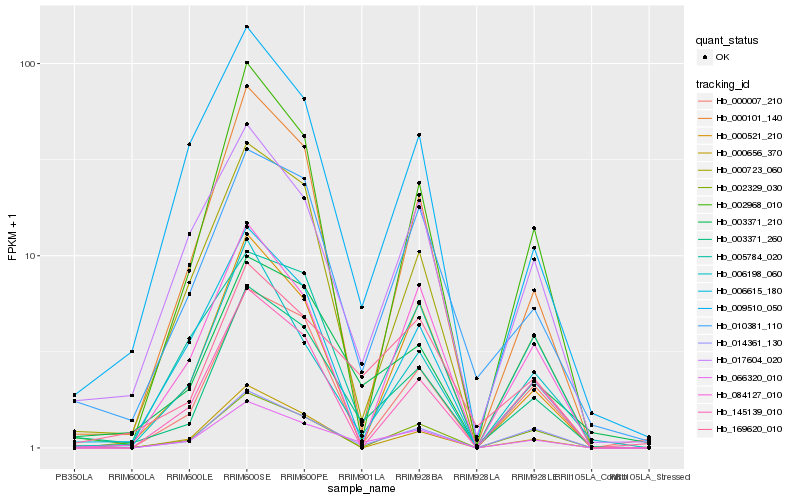
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066320_010 |
0.0 |
- |
- |
hypothetical protein VITISV_027639 [Vitis vinifera] |
| 2 |
Hb_014361_130 |
0.1153944438 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 3 |
Hb_003371_260 |
0.1261810046 |
- |
- |
hypothetical protein EUGRSUZ_B03661 [Eucalyptus grandis] |
| 4 |
Hb_003371_210 |
0.1468867886 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_169620_010 |
0.1485037845 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 6 |
Hb_000101_140 |
0.149642044 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_017604_020 |
0.1516806954 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 8 |
Hb_002968_010 |
0.1532486998 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 9 |
Hb_000723_060 |
0.1534050685 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 10 |
Hb_006615_180 |
0.1541421321 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 11 |
Hb_002329_030 |
0.155122657 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_009510_050 |
0.1564607294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005784_020 |
0.1571952966 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 14 |
Hb_000521_210 |
0.1573824845 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 15 |
Hb_084127_010 |
0.1580147977 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000656_370 |
0.1631259368 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_006198_060 |
0.1649855852 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 18 |
Hb_010381_110 |
0.1651434821 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 19 |
Hb_000007_210 |
0.1663730195 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 20 |
Hb_145139_010 |
0.1668167079 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |