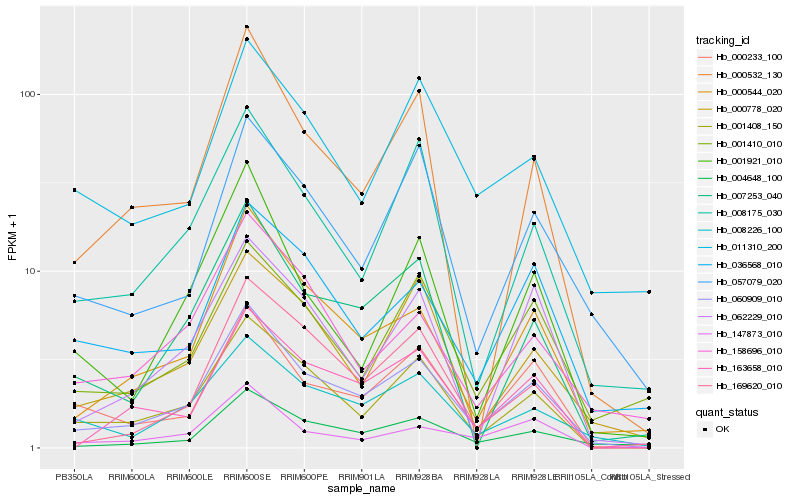
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060909_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 2 |
Hb_001408_150 |
0.1080043239 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000544_020 |
0.1133677714 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_000233_100 |
0.1201168565 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_004648_100 |
0.1243360874 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 6 |
Hb_008226_100 |
0.1250262061 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_000532_130 |
0.132159727 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 8 |
Hb_007253_040 |
0.1381664743 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 9 |
Hb_163658_010 |
0.1417424869 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_008175_030 |
0.1425764492 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_057079_020 |
0.1469635293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000778_020 |
0.1476895929 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 13 |
Hb_169620_010 |
0.1492615559 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 14 |
Hb_062229_010 |
0.1539277772 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_036568_010 |
0.1541873651 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 16 |
Hb_147873_010 |
0.1547912705 |
- |
- |
PREDICTED: uncharacterized protein LOC102595311 [Solanum tuberosum] |
| 17 |
Hb_001410_010 |
0.1562199348 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 18 |
Hb_011310_200 |
0.1562227568 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 19 |
Hb_001921_010 |
0.1566742905 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_158696_010 |
0.1579391415 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |