| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
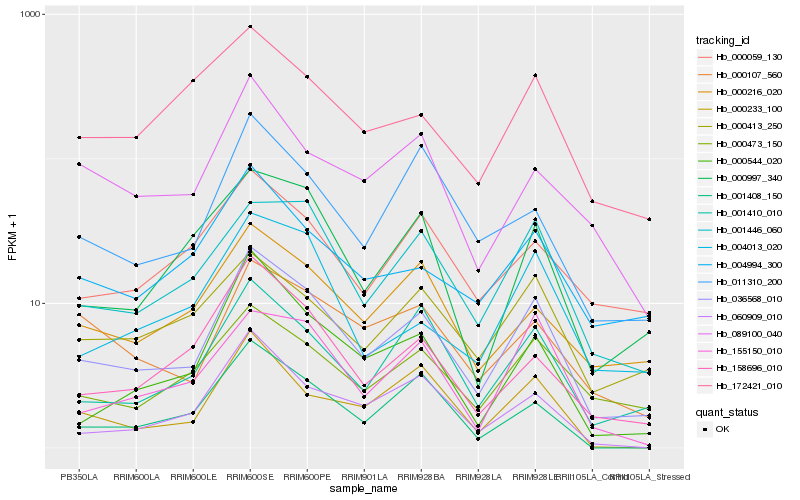
| 1 |
Hb_036568_010 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 2 |
Hb_001410_010 |
0.1331595468 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 3 |
Hb_001446_060 |
0.1367652682 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000473_150 |
0.1418289889 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000997_340 |
0.1478683701 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000544_020 |
0.14939259 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 7 |
Hb_004994_300 |
0.149490381 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 8 |
Hb_158696_010 |
0.1499757078 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 9 |
Hb_000216_020 |
0.1506726654 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 10 |
Hb_004013_020 |
0.1534284259 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 11 |
Hb_060909_010 |
0.1541873651 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 12 |
Hb_155150_010 |
0.1563521551 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_000107_560 |
0.1566154728 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 14 |
Hb_011310_200 |
0.1573590314 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 15 |
Hb_000233_100 |
0.1583307628 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_000413_250 |
0.159168903 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 17 |
Hb_001408_150 |
0.1603708237 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_130 |
0.1614708339 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 19 |
Hb_172421_010 |
0.1646556106 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 20 |
Hb_089100_040 |
0.1649073551 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |