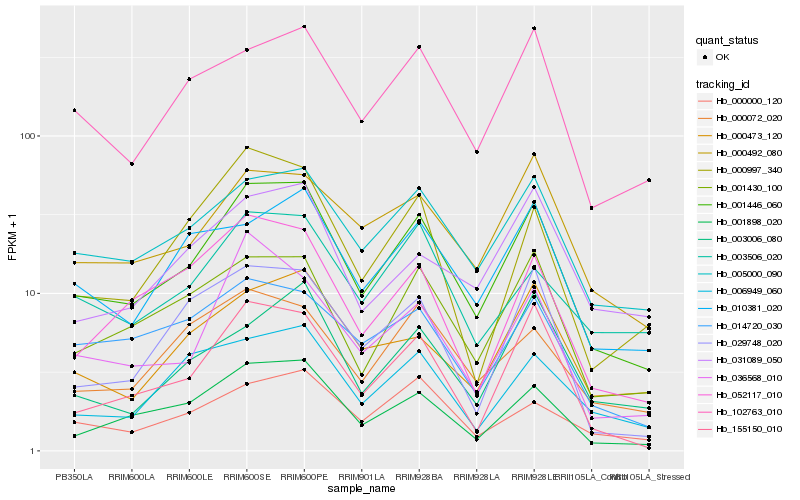
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001446_060 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001898_020 |
0.098756291 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 3 |
Hb_005000_090 |
0.1017918366 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 4 |
Hb_001430_100 |
0.1097497047 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_155150_010 |
0.1132450761 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_102763_010 |
0.1166175711 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 7 |
Hb_000492_080 |
0.1251297435 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 8 |
Hb_000000_120 |
0.1272848797 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 9 |
Hb_010381_020 |
0.1284889128 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 10 |
Hb_000473_120 |
0.1294431465 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 11 |
Hb_052117_010 |
0.129990335 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000997_340 |
0.1326557495 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_006949_060 |
0.1332869213 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 14 |
Hb_029748_020 |
0.1352488031 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000072_020 |
0.1355963071 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 16 |
Hb_036568_010 |
0.1367652682 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 17 |
Hb_003506_020 |
0.1383330424 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 18 |
Hb_014720_030 |
0.1385060734 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 19 |
Hb_003006_080 |
0.1389106934 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_031089_050 |
0.138935764 |
- |
- |
kinase, putative [Ricinus communis] |