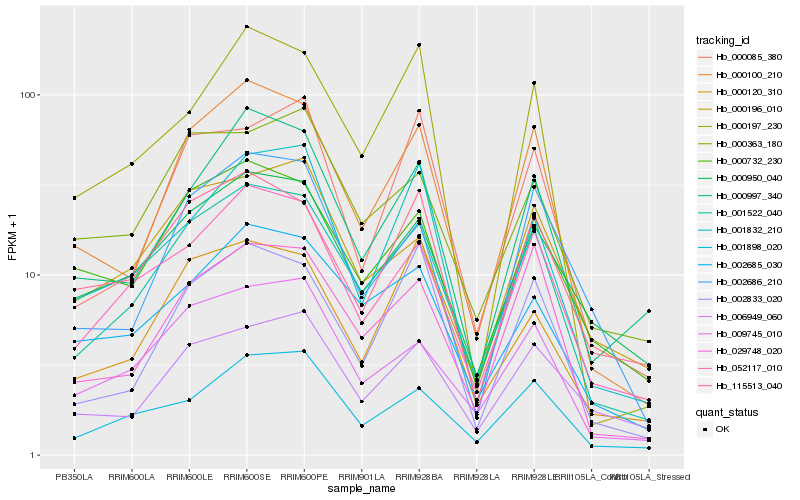
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001522_040 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_052117_010 |
0.0800949493 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000100_210 |
0.0879644289 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 4 |
Hb_009745_010 |
0.0937705678 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 5 |
Hb_001898_020 |
0.0966574507 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 6 |
Hb_029748_020 |
0.0988615792 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_002685_030 |
0.1056368257 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 8 |
Hb_000950_040 |
0.1069746301 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 9 |
Hb_000732_230 |
0.1093675377 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000997_340 |
0.1129722435 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000196_010 |
0.1192176939 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 12 |
Hb_000363_180 |
0.1204839293 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 13 |
Hb_115513_040 |
0.1213376511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002833_020 |
0.1220283762 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_002686_210 |
0.1250873401 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_006949_060 |
0.127067751 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 17 |
Hb_001832_210 |
0.1314247187 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 18 |
Hb_000120_310 |
0.1321246222 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_000085_380 |
0.1348477518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000197_230 |
0.135026424 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |