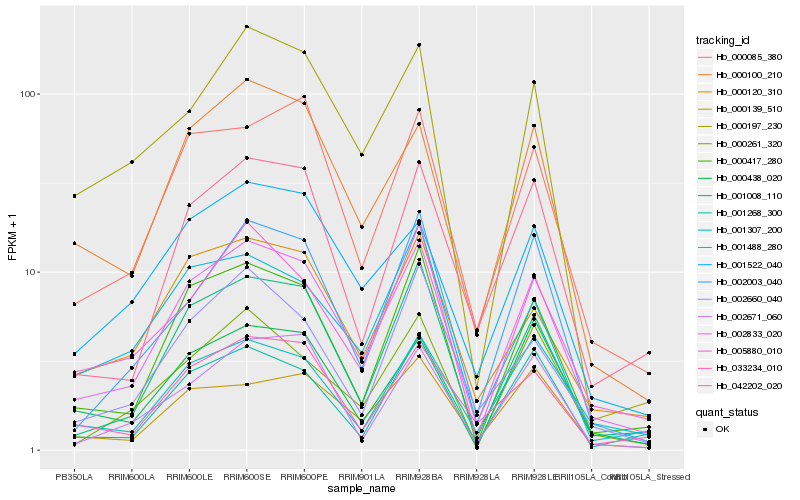
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002833_020 |
0.0 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_001008_110 |
0.0835592725 |
- |
- |
- |
| 3 |
Hb_000417_280 |
0.0872463438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000100_210 |
0.1036461397 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 5 |
Hb_000085_380 |
0.10692579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000120_310 |
0.1103743535 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 7 |
Hb_042202_020 |
0.1104058946 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 8 |
Hb_005880_010 |
0.1120697864 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 9 |
Hb_033234_010 |
0.1140530956 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 10 |
Hb_002003_040 |
0.1147455892 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000261_320 |
0.1170181163 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000139_510 |
0.1208713807 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001522_040 |
0.1220283762 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_002660_040 |
0.1232731396 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001268_300 |
0.1236827994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000197_230 |
0.1237697659 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 17 |
Hb_001488_280 |
0.1270222735 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 18 |
Hb_001307_200 |
0.1282119947 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 19 |
Hb_000438_020 |
0.1307124812 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 20 |
Hb_002671_060 |
0.1316256544 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |