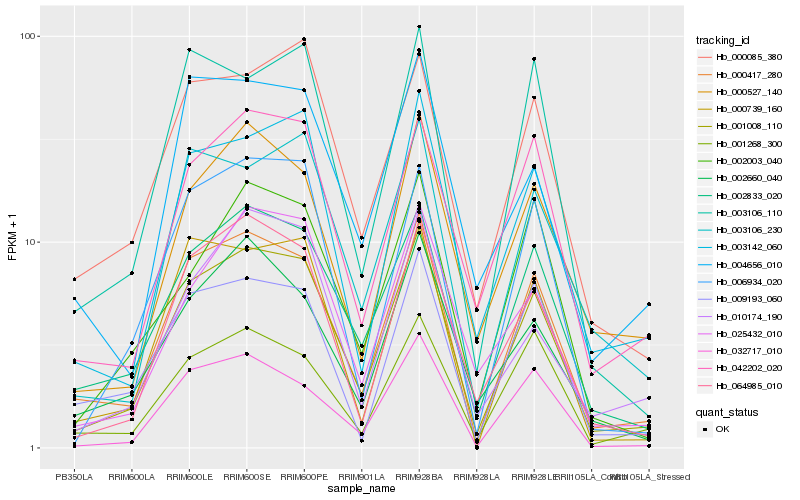
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_000417_280 |
0.0615777396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002833_020 |
0.0835592725 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_064985_010 |
0.0983650036 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 5 |
Hb_003142_060 |
0.1056267841 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_032717_010 |
0.1095104782 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_042202_020 |
0.1099623586 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 8 |
Hb_006934_020 |
0.1117931808 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 9 |
Hb_000085_380 |
0.1136742747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000527_140 |
0.1137606779 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 11 |
Hb_000739_160 |
0.1145718941 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003106_230 |
0.1158595376 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 13 |
Hb_025432_010 |
0.1169480021 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 14 |
Hb_001268_300 |
0.1218041509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002660_040 |
0.1247770118 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010174_190 |
0.1264919564 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 17 |
Hb_002003_040 |
0.1265563487 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003106_110 |
0.1271110473 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_009193_060 |
0.1274604514 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 20 |
Hb_004656_010 |
0.129114445 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |