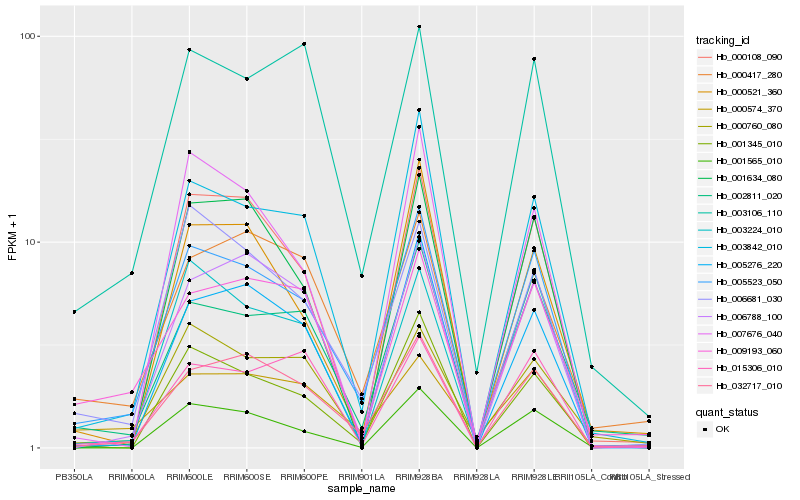
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005523_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 2 |
Hb_032717_010 |
0.1020028998 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_003842_010 |
0.1137767704 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_002811_020 |
0.1179014651 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 5 |
Hb_006681_030 |
0.1264469857 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_001345_010 |
0.129834124 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 7 |
Hb_003224_010 |
0.1385754271 |
- |
- |
- |
| 8 |
Hb_005276_220 |
0.1427998847 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 9 |
Hb_006788_100 |
0.1428387436 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003106_110 |
0.1429300138 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000108_090 |
0.1461116579 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 12 |
Hb_001634_080 |
0.1492021077 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_001565_010 |
0.1520296829 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 14 |
Hb_007676_040 |
0.153849369 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 15 |
Hb_000417_280 |
0.1540718943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_015306_010 |
0.1544439907 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 17 |
Hb_000760_080 |
0.1558610968 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 18 |
Hb_000521_360 |
0.1568190902 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_009193_060 |
0.1570395705 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 20 |
Hb_000574_370 |
0.1579763058 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |