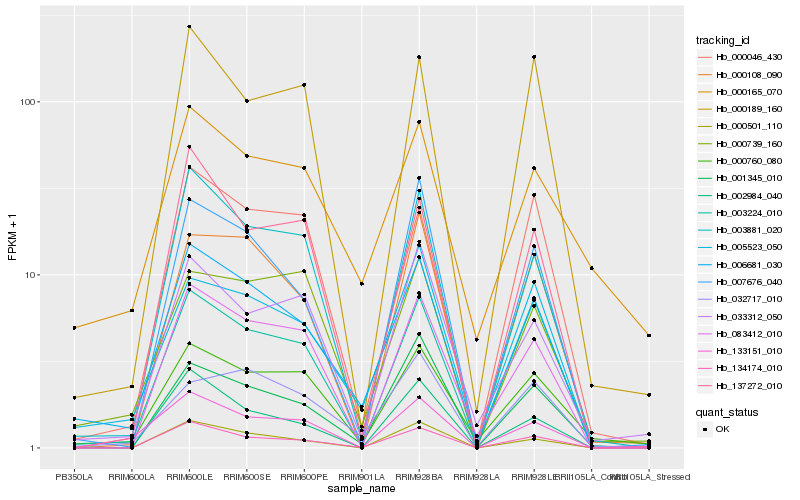
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006681_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_005523_050 |
0.1264469857 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 3 |
Hb_003881_020 |
0.131962506 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 4 |
Hb_000760_080 |
0.1411906322 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 5 |
Hb_003224_010 |
0.1446892836 |
- |
- |
- |
| 6 |
Hb_134174_010 |
0.1487182762 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_007676_040 |
0.1514309708 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 8 |
Hb_083412_010 |
0.1524713218 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_000501_110 |
0.1530423523 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 10 |
Hb_000108_090 |
0.1581499493 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 11 |
Hb_032717_010 |
0.1588582025 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000046_430 |
0.1596822107 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 13 |
Hb_002984_040 |
0.1620483031 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001345_010 |
0.1624373456 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 15 |
Hb_133151_010 |
0.1654695491 |
- |
- |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 16 |
Hb_000165_070 |
0.165675988 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 17 |
Hb_033312_050 |
0.1661006225 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 18 |
Hb_137272_010 |
0.166145094 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 19 |
Hb_000739_160 |
0.1670692209 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000189_160 |
0.1672993332 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |